- Title
-
When Bigger Is Better: 3D RNA Profiling of the Developing Head in the Catshark Scyliorhinus canicula
- Authors
- Mayeur, H., Lanoizelet, M., Quillien, A., Menuet, A., Michel, L., Martin, K.J., Dejean, S., Blader, P., Mazan, S., Lagadec, R.
- Source
- Full text @ Front Cell Dev Biol
|
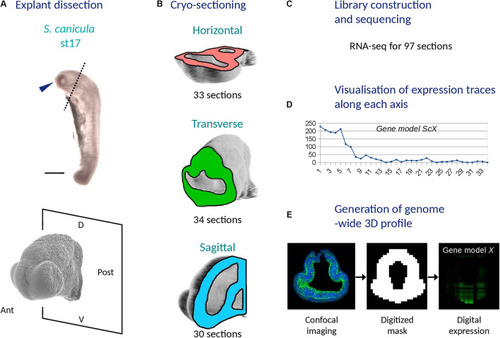
Main experimental steps used to generate a 3D RNA profile of the catshark embryonic head (stage 17). |
|
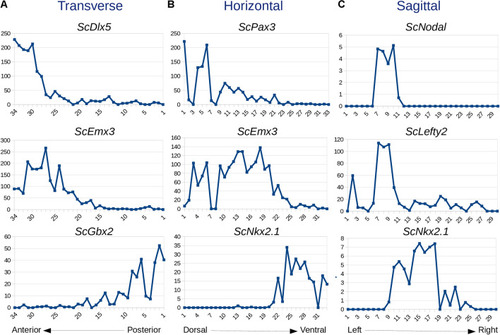
Expression traces for selected genes along transverse |
|
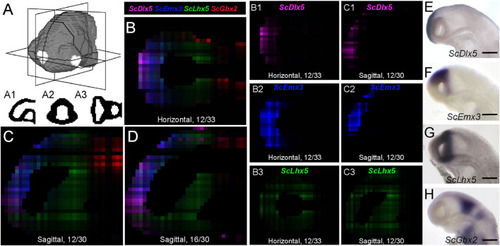
Digital profiles reproduce regional patterns along AP axis. |
|
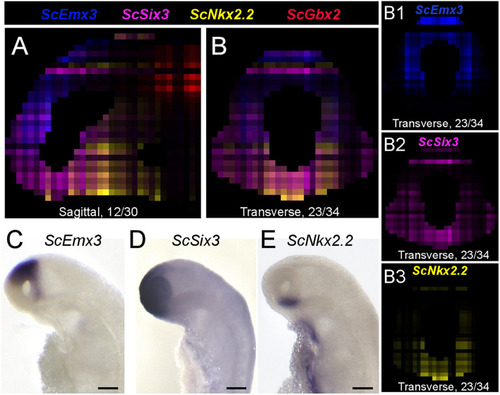
Digital profiles reproduce regional patterns along DV axis. |
|
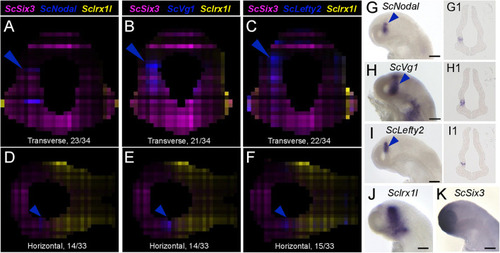
Figure 5. Nodal signaling components exhibit left enriched profiles. (A–F) Digital transverse (A–C) and horizontal (D–F) sections showing merged profiles of ScSix3 (magenta), ScIrx1l (yellow), and in blue ScNodal (A,D), ScVg1 (B,E), ScLefty2 (C,F). Sections are numbered from posterior to anterior in panels (A–C) and from dorsal to ventral in panels (D–F). (G–K) Left lateral views of stage 17 catshark embryonic heads after whole-mount ISH with probes for ScNodal (G), ScVg1 (H), ScLefty2 (I), ScIrx1l (J), and ScSix3 (K). (G1–I1) Horizontal sections of the brain of stage 17 catshark embryos after hybridization with probes for ScNodal (G1), ScVg1 (H1), ScLefty2 (I1). Blue arrowheads point to ScNodal, ScVg1, and ScLefty2 left-restricted digital or ISH signals in the dorsal diencephalon. Scale bars = 200 μm. |
|
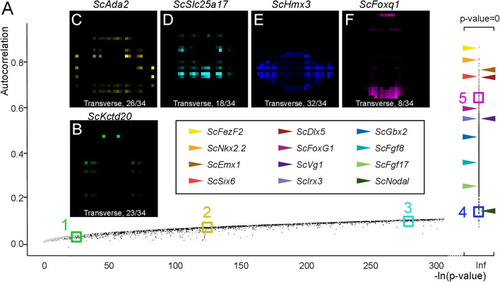
Autocorrelation as an indicator of profile regionalization. |
|
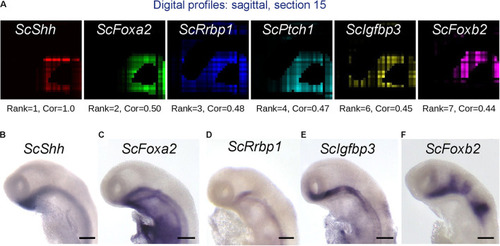
Figure 7. Identification of genes harboring expression correlation with ScShh. (A) Digital profiles on sagittal section number 15 (out of 30, from left to right) of a selection of genes showing a high expression correlation to ScShh (red), ScFoxA2 (green), ScRrbp1 (blue), ScPtch1 (cyan), ScIgfbp3 (yellow), and ScFoxb2 (purple). The rank of each gene in the list of gene models ordered by decreasing correlation to ScShh and the correlation value (Cor) are indicated below each section. (B–F) Left lateral views of stage 17 catshark embryonic heads after ISH using probes for ScShh (B), ScFoxA2 (C), ScRrbp1 (D), ScIgfbp3 (E), and ScFoxb2 (F). Scale bars = 200 μm. |