- Title
-
Metabolic Alterations in Preneoplastic Development Revealed by Untargeted Metabolomic Analysis
- Authors
- Myllymäki, H., Astorga Johansson, J., Grados Porro, E., Elliot, A., Moses, T., Feng, Y.
- Source
- Full text @ Front Cell Dev Biol
|
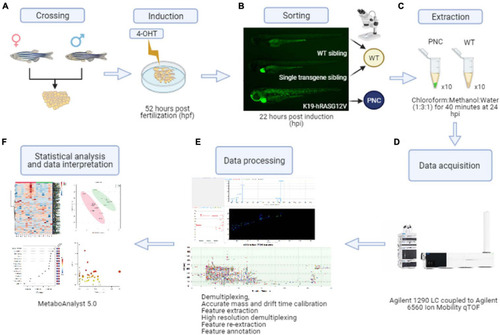
Preneoplastic cell (PNC) induction and schematic workflow for untargeted metabolomics from zebrafish larval skin. |
|
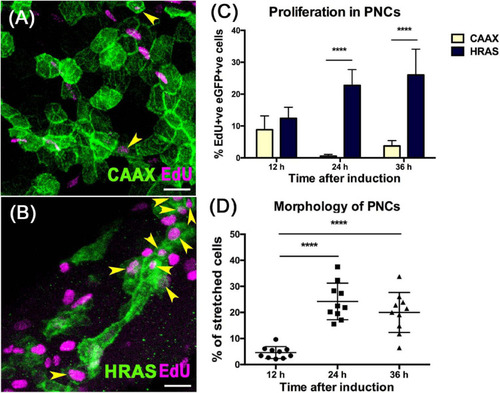
HRASG12V expressing PNCs show increased proliferation and altered morphology from 24 hpi. PHENOTYPE:
|
|
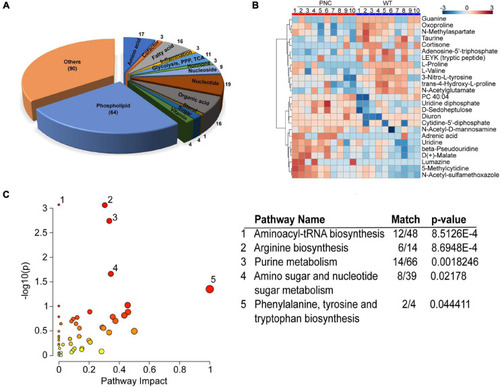
Metabolites extracted from zebrafish larval skin tissue. |
|
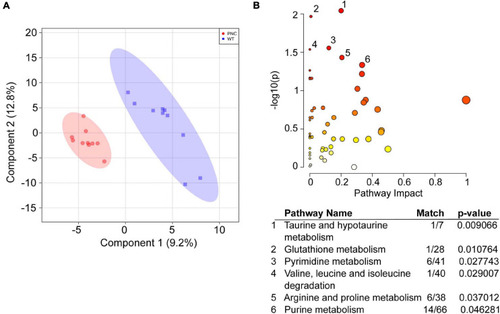
Metabolic pathways significantly altered upon HRASG12V induction in PNCs. |
|
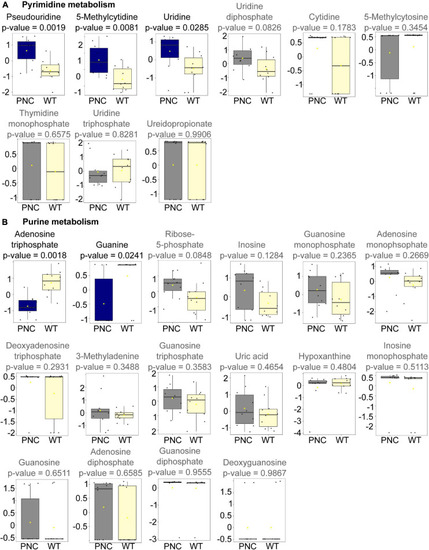
Metabolites significantly altered upon HRASG12V induction in PNCs. Box and whisker plots generated using MetaboAnalyst 5.0 online platform for individual metabolites in pyrimidine PHENOTYPE:
|
|
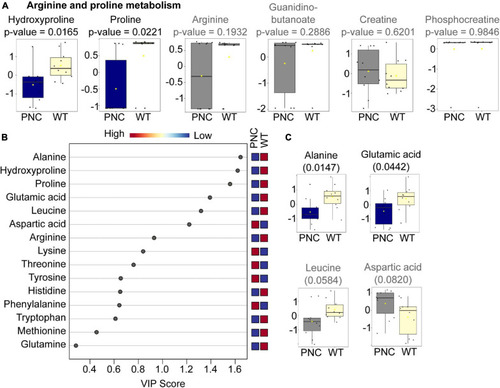
Amino acids significantly altered upon HRASG12V induction in PNCs. PHENOTYPE:
|
|
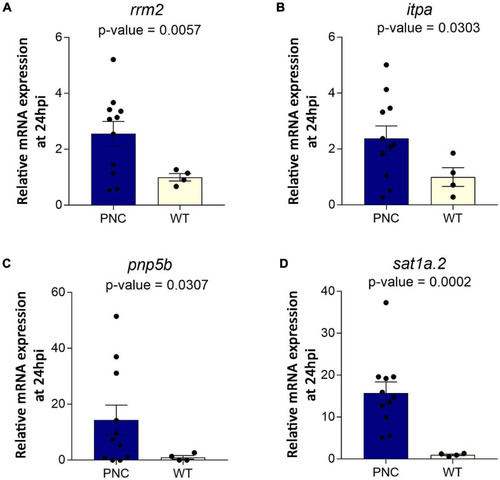
Upregulation of genes encoding metabolic enzymes at 24 h post HRAS EXPRESSION / LABELING:
PHENOTYPE:
|