FIGURE 3
- ID
- ZDB-FIG-210822-18
- Publication
- Myllymäki et al., 2021 - Metabolic Alterations in Preneoplastic Development Revealed by Untargeted Metabolomic Analysis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
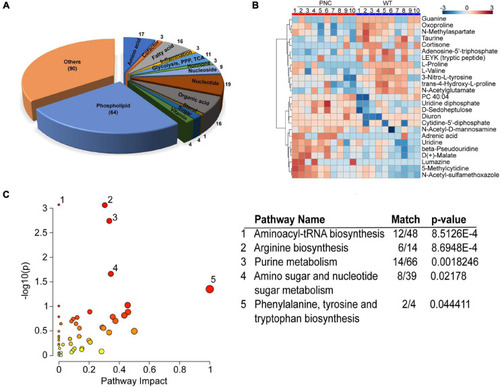
Metabolites extracted from zebrafish larval skin tissue. |