- Title
-
Syntabulin, a motor protein linker, controls dorsal determination
- Authors
- Nojima, H., Rothhämel, S., Shimizu, T., Kim, C.H., Yonemura, S., Marlow, F.L., and Hibi, M.
- Source
- Full text @ Development
|
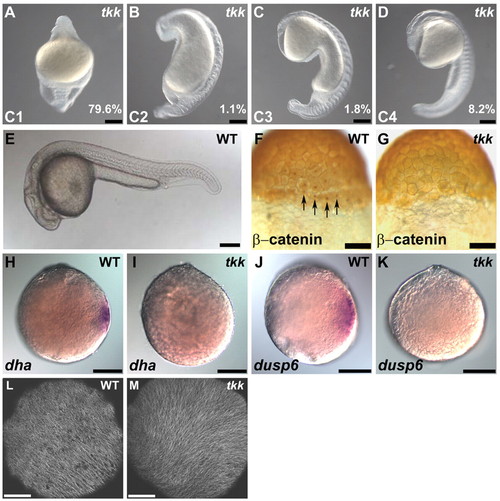
Maternal-effect tokkaebi (tkk) mutant zebrafish embryos show defects in dorsal determination. (A-E) Phenotypes of tkk embryos (A-D) versus wild type (WT; E) at 24 hpf. Lateral views with dorsal to the right (A-D) or to the top (E). Embryos were classified into four classes (C1-4) based on their ventralized phenotypes. C1 embryos displayed a completely ventralized phenotype. C2 embryos were similar to C1 embryos but had somites in the trunk region. C3 embryos had somites and spinal cord in the trunk. C4 embryos had somites, spinal cord and hindbrain, but lacked notochord or neuroectoderm anterior to the midbrain. The percentage of tkk embryos from typical young females that showed each phenotype is indicated. (F,G) Immunostaining of β-catenin at the 256-cell stage in wild-type (F; 88%, n=26) and tkk (G; 11%, n=35) embryos. Dorsal views. Arrows indicate nuclear accumulation of β-catenin in dorsal blastomeres. (H,I) Expression of dha at sphere stage (4 hpf) in wild-type (H; 100%, n=36) and tkk (I; 8%, n=40) embryos. (J,K) Expression of dusp6 at sphere stage in wild-type (J; 100%, n=29) and tkk (K; 6%, n=36) embryos. Animal pole views. (L,M) Immunostaining with anti-β-tubulin antibody. Wild-type (L; 86%, n=14) and tkk (M; 81%, n=16) embryos at 20 mpf. Vegetal pole views. Scale bars: 200 μm in A-E,H-K; 140 μm in F,G; 20 μm in L,M. |
|
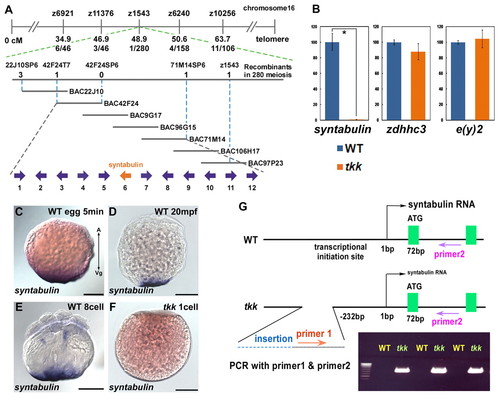
syntabulin is responsible for the tkk mutant phenotype. (A) Linkage mapping. The zebrafish tkk locus was initially mapped near the SSLP marker z1543. Seven BAC clones were used to construct a physical map. Linkage analysis (numbers indicate recombinants per meiosis), sequence analysis of BAC clones and database searches revealed that there were 12 candidate genes in the chromosomal region. (B) Quantification of syntabulin (gene 6 in A) expression in wild-type and tkk embryos (expression in wild type taken as 100; *, P<0.01; the average ±s.d. is shown). zdhhc3 (zinc-finger, DHHC-type containing 3, gene 3) and e(y)2 (enhancer of yellow gene, gene 8) are located in the same chromosomal region as syntabulin and were used as controls. The expression of other genes in this chromosomal region is shown in Table S1 in the supplementary material. (C-F) Expression and localization of syntabulin RNA in an unfertilized wild-type egg (5 minutes after activation, C), in a 1-cell stage wild-type embryo (20 mpf, D), an 8-cell stage wild-type embryo (E), and in a 1-cell stage tkk embryo (F). Lateral views; A, animal pole; Vg, vegetal pole. (G) Molecular nature of the syntabulintkk4 allele. PCR detection of the insertion in wild-type and tkk embryos (n=3 each). Scale bars: 200 μm. |
|
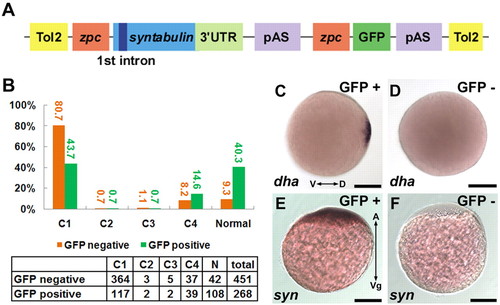
Maternal expression of syntabulin rescues tkk mutants. (A) Structure of the rescue plasmid. Tol2, inverted repeats of Tol2 transposon; zpc, promoter of zone pellucida protein C; 3′ UTR, 3′ untranslated region; pAS, polyadenylation signal. (B) Rescue by syntabulin cDNA transgene. The numbers of transgenic (green) and non-transgenic (orange) zebrafish embryos showing each phenotype are tabulated, and the percentages are shown in the bar chart. (C,D) Expression of dha in tkk (D; 8%, n=25) and transgene-rescued (C, GFP+; 48%, n=33) embryos at the late blastula stage. (E,F) Localization of syntabulin at the 1-cell stage in a transgene-rescued (E; 67%, n=27) versus control (F; 100%, n=23) embryo. Scale bars: 200 μm. |
|
Exon-intron structure of syntabulin is required for vegetal localization and proper dorsal determination. (A-E) The 3′ UTR is not sufficient for vegetal localization of syntabulin. (A) Structure of the transgene plasmid. (B-E) Localization of GFP mRNA in transgenic (B,D) or non-transgenic (C,E) tkk (D,E) or wild-type (B,C) zebrafish embryos at the 1-cell stage (20 mpf). (F,G) Genomic structure of syntabulin (F) and the rescue plasmid that contains the exon-intron structure of syntabulin. 2A-peptide, 2A peptide of PTV1. (H) Rescue by genomic DNA. The numbers (tabulated) and percentages (bar chart) of transgenic (green) and non-transgenic (orange) embryos showing each phenotype. (I,J) Expression of dha in tkk (J; 9%, n=32) and rescued (I; 66%, n=33) embryos at the late blastula stage. (K,L) Localization of syntabulin at the 1-cell stage in a rescued (K; 86%, n=35) versus control (L; 100%, n=31) embryo. Shown are animal pole views (I,J) and lateral views (A-E,K,L). Vegetal pole localization is marked by an asterisk (K). Scale bars: 200 μm. |
|
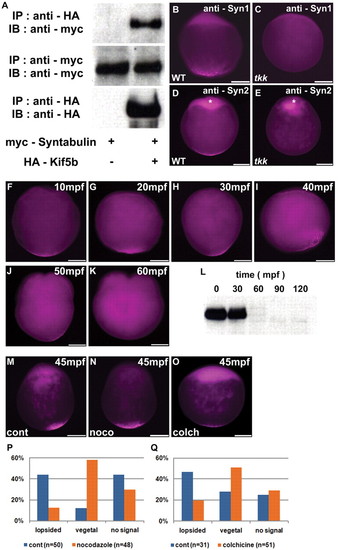
Microtubule-dependent transportation of Syntabulin. (A) Interaction between Syntabulin and Kif5b. HEK293T cells were transfected with expression vectors of Myc-tagged Syntabulin and HA-tagged Kif5b. IP, immunoprecipitation; IB, immunoblotting. (B-E) Detection of Syntabulin with anti-Syntabulin monoclonal antibodies (anti-Syn1, anti-Syn2) in wild-type and tkk zebrafish embryos at early zygote stage (10 mpf). Expression of Syntabulin at the animal pole (asterisks in D,E) is non-specific as it is detected in tkk embryos, which lack significant amounts of Syntabulin (see Fig. S8C in the supplementary material). (F-K) Time course of Syntabulin localization in the wild type. Immunostaining with anti-Syn1. (L) Immunoblotting of anti-Syntabulin (anti-Syn2) immunoprecipitates. (M-O) Localization of Syntabulin at 45 mpf in wild-type embryos treated with 1 μg/ml nocodazole (N) or 0.4 mg/ml colchicine (O) just after fertilization, versus untreated control (M). (P,Q) Quantification of Syntabulin localization at 45 mpf showing lopsided or vegetal pole localization, or undetectable expression in embryos treated with nocodazole (P) or colchicine (Q), versus control (P,Q). Shown are lateral views with animal pole to the top. Scale bars: 200 μm. |
|
Balbiani body-mediated localization of syntabulin at the vegetal pole. (A-D) Localization of syntabulin RNA in wild-type and bucky ball mutant (bucp106 and bucp43) stage Ib (STI/Ib) zebrafish oocytes. Staining by whole-mount in situ hybridization with sense (A, control) or antisense (B-D) riboprobes. In wild-type oocytes, syntabulin RNA localizes to the Balbiani body (A,B; 84%, n=104). In bucp106 (C; 97%, n=87 oocytes from three females) and bucp43 (D; 100%, n=36 from one female) mutant oocytes, syntabulin is not spatially restricted or localized. (E,F) Localization of syntabulin RNA. In wild-type oocytes (E), syntabulin localizes to the Balbiani body, whereas syntabulin is detected throughout the cytoplasm in bucp106 mutants (F; 100%, n=27 from one female). (G) Quantification of syntabulin expression in wild-type and buc oocytes. Although syntabulin is not localized in buc mutants, the transcripts are present in early and late stage oocytes. (H-J) Localization of syntabulin RNA in wild-type and buc activated eggs. In activated eggs from wild type, syntabulin shows polarized localization (H; 100% n=37 from two females). In activated eggs from bucp106 (I; 100%, n=44 from three females) and bucp43 (J; 100%, n=8 from one female) mutants, syntabulin is detected around the circumference. Arrows indicate localization of syntabulin at the vegetal pole in wild type and its circumferential expansion in buc eggs. nuc, nucleus; Bb, Balbiani body. Scale bars: 50 μm. |
|
Overview of organizer induction in zebrafish. (A) Schematic representation of dorsal organizer formation. (B) Evidence for vegetal localization of the dorsal determinants and involvement of microtubules in dorsal determination. |
|
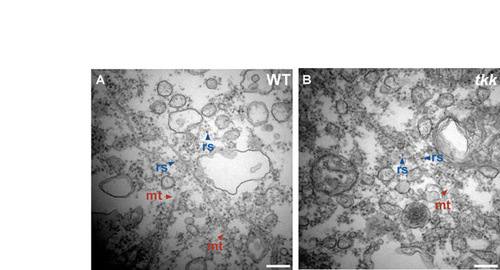
Organelles at the vegetal pole. (A,B) Electron microscopy analysis. Wild-type (A) and tkk (B) embryos at 20 mpf. Vegetal pole regions. mt, microtubule; rs, ribosome. Scale bars: 0.2 μm. |
|
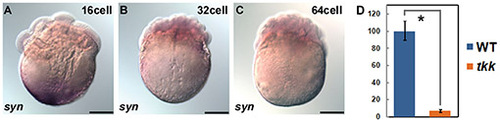
Localization and expression of syntabulin RNA. (A-C) Localization of syntabulin RNA in wild-type (WT) embryos at 16-cell stage (A), 32-cell stage (B) and 64-cell stage (C). syntabulin was detected at the vegetal pole at the 16-cell stage, and weakly detected in blastomeres at the 32- and 64-cell stages. (D) Quantification (qRT-PCR) of syntabulin expression in wild-type and tkk embryos at 24 hpf (*, P<0.01). Scale bars: 200 μm. EXPRESSION / LABELING:
|
|
Detection of the insertion by Southern blotting. (A) Schematic representation of the zebrafish syntabulin gene structure and location of the probe used for Southern blotting analysis. (B) Southern blot. Genomic DNA (10 μg) was isolated from four wild-type and four tkkrk4/rk4 adult fish and digested with EcoRI, followed by Southern hybridization with the probe described in A. A larger than 10 kb band was detected in the wild-type genome, whereas a <7 kb band was detected in the tkk genome (asterisks), indicating alteration of the 5′ syntabulin genomic sequence. |
|
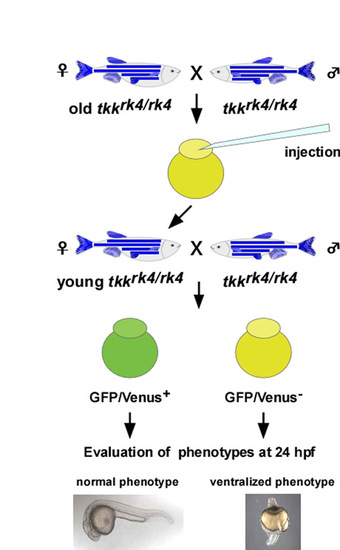
Rescue procedure. Relatively old (older than 6 months) tkkrk4/rk4 female fish were crossed with tkkrk4/rk4 male fish to produce mildly affected or non-phenotypic offspring. Rescue plasmid (Fig. 3A, Fig. 5G) and Tol2 transposase RNA were injected into these 1-cell stage embryos. The tkkrk4/rk4 female fish that were positive for the transgenes were raised to adulthood and crossed with tkkrk4/rk4 male fish. GFP/Venus+ (transgenic) and GFP/Venus- (non-transgenic) embryos were sorted after fertilization and their phenotypes were classified at 24 hpf (Fig. 3B, Fig. 5H). |
|
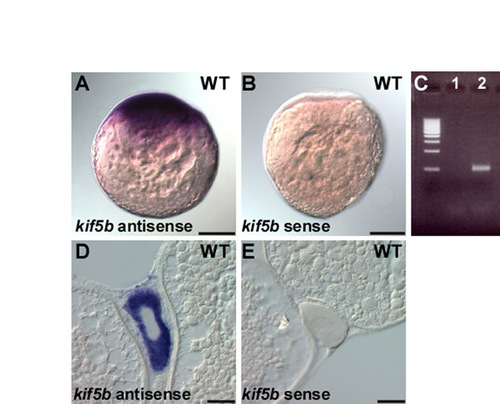
Maternal expression of kif5b RNA. (A,B) Maternal expression of kif5b. One-cell stage embryos (20 mpf) were fixed and hybridized with antisense (A) or sense control (B) probe. The kif5b probe was produced by RT-PCR. (C) RT-PCR. cDNA was generated from 1-cell stage embryos with (lane 2) or without (lane 1) reverse transcriptase, and a kif5b cDNA fragment was amplified by RT-PCR. Primers used for the kif5b detection are 5′-GATTCGGTAACCGGGGGAAGATGG-3′ and 5′-TCCACATTGACACACACAGTATTC-3′. (D,E) kif5b mRNA is ubiquitously distributed in a stage I oocyte as revealed by in situ hybridization of wild-type ovary sections with antisense (D) versus sense (E) probes. Scale bars: 200 μm in A,B; 50 μm in D,E. |
|
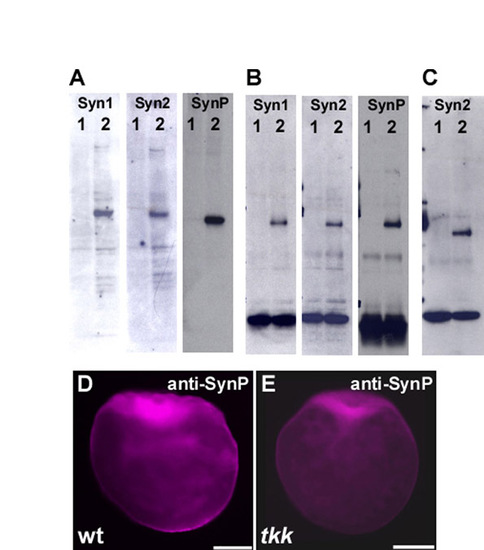
Evaluation of anti-Syntabulin monoclonal and polyclonal antibodies. (A) Immunoblot detection of Syntabulin in lysates of HEK293T cells that were transfected with zebrafish Syntabulin expression plasmid (lane 2) or not transfected (lane 1). Ten microliters of 1 ml lysates of transfected HEK293T cells in a 6-cm dish were used. (B) Lysates from embryos just after fertilization (lane 2) or 60 mpf (lane1) were immunoprecipitated and immunoblotted with anti-Syntabulin antibodies. (C) Lysates from tkk (lane 1) or wild-type (lane 2) embryos just after fertilization were immunoprecipitated and immunoblotted with anti-Syntabulin (anti-Syn2) antibodies. (D,E) Immunostaining of wild-type and tkk embryos (fixed just after fertilization) with polyclonal anti-Syntabulin antibody (anti-SynP). Mouse monoclonal antibodies, anti-Syn1 and anti-Syn2, were raised against GST fusion proteins containing amino acids 14-94 and 102-271, respectively (see Fig. S2). Rabbit polyclonal antibody, anti-SynP, was raised against His-tagged protein containing amino acids 1-120 of Syntabulin. Anti-SynP was affinity purified with the antigen-conjugated column. Scale bars: 200 μm. |
|
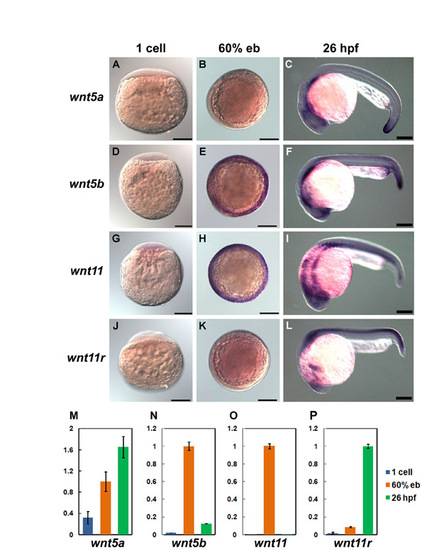
Expression of wnt5a/5b and wnt11/11r. (A-L) Whole-mount in situ hybridization. Lateral views at 1-cell stage (A,D,G,J), animal pole views with dorsal to the right at 60% epiboly (B,E,H,K), and lateral views with anterior to the left (C,F,I,L). The detection of wnt5b (Rauch et al., 1997), wnt11 (Heisenberg et al., 2000) and wnt11r (Matsui et al., 2005) was described previously. The wnt5a probe was prepared by RT-PCR. (M-P) Quantification by qRT-PCR. Expression levels of wnt5a/5b and wnt11 at 60% epiboly, and wnt11r at 26 hpf, were taken as 100. Primers: wnt5a, 5′-GAACTGCAGCACCGTCGACAACTC-3′ and 5′-GAACTGCAGCACCGTCGACAACTC-3′; wnt5b, 5′-GCCTATCTCAGGGTCAGAGGAA-3′ and 5′-CGCCCCCTCTCCAATATAAAC-3′; wnt11, 5′-CACCCCTGACCTGGCCAAAGGGAC-3′ and 5′-GGAGATGGTGCTGATGTCTTGA-3′; wnt11r, 5′-TCTCACGCCAACAAGCTCAT-3′ and 5′-CCAGAAACACCGTGGCATT-3′. Scale bars: 200 μm. |
|
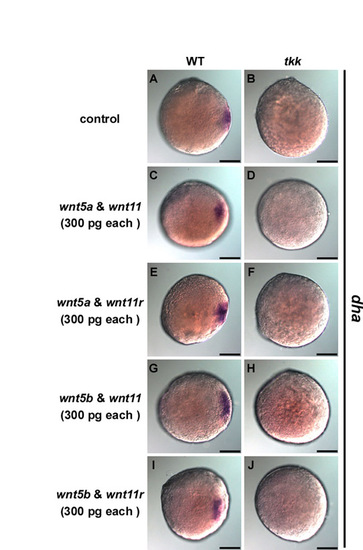
Overexpression of wnt5a/5b and wnt11/11r does not rescue tkk or dorsalize the tkk embryos. (A-J) wnt5a, wnt5b, wnt11 and wnt11r RNAs (300 pg) were injected into 1-cell stage wild-type (A,C,E,G,I) and tkk (B,D,F,H,J) embryos just after fertilization in the indicated combinations. Expression of dharma (dha) was examined at the late blastula stage (4 hpf, sphere stage). Animal pole views with dorsal to the left. Scale bars: 200 μm. |