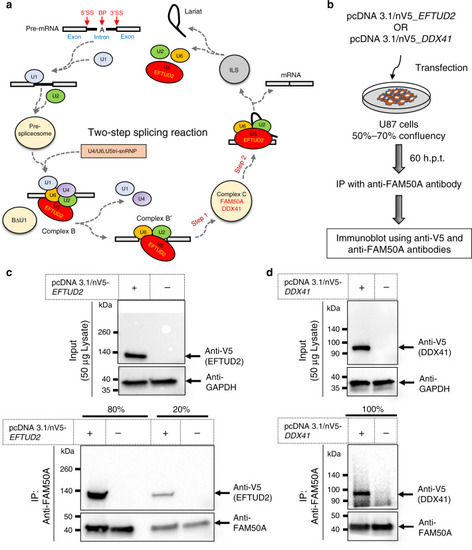
- Title
-
Mutations in FAM50A suggest that Armfield XLID syndrome is a spliceosomopathy
- Authors
- Lee, Y.R., Khan, K., Armfield-Uhas, K., Srikanth, S., Thompson, N.A., Pardo, M., Yu, L., Norris, J.W., Peng, Y., Gripp, K.W., Aleck, K.A., Li, C., Spence, E., Choi, T.I., Kwon, S.J., Park, H.M., Yu, D., Do Heo, W., Mooney, M.R., Baig, S.M., Wentzensen, I.M., Telegrafi, A., McWalter, K., Moreland, T., Roadhouse, C., Ramsey, K., Lyons, M.J., Skinner, C., Alexov, E., Katsanis, N., Stevenson, R.E., Choudhary, J.S., Adams, D.J., Kim, C.H., Davis, E.E., Schwartz, C.E.
- Source
- Full text @ Nat. Commun.
|
|
|
EXPRESSION / LABELING:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
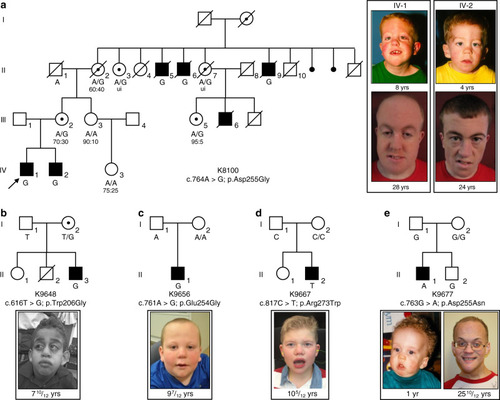
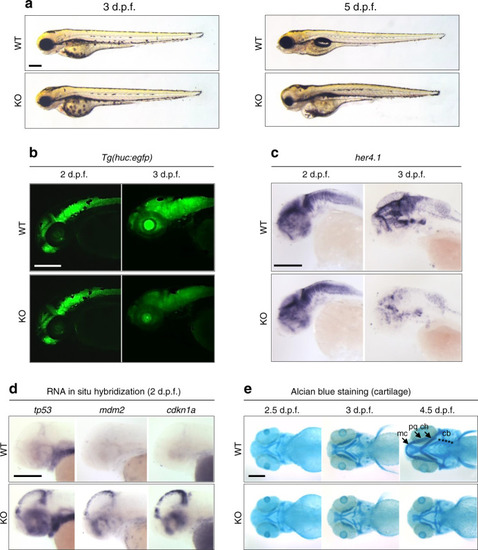
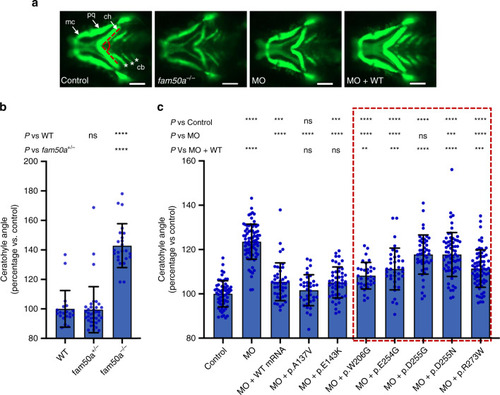
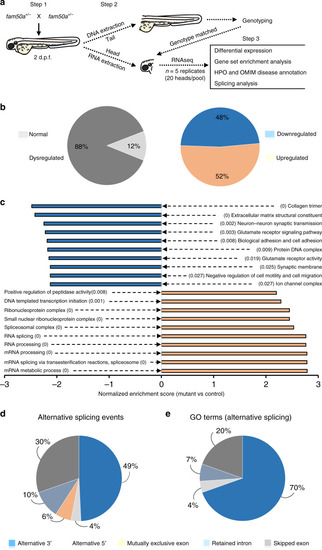
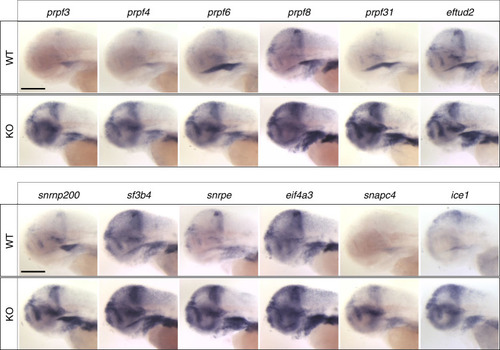
Representative lateral images of whole-mount in situ hybridization performed on 3 day post-fertilization larvae validate RNA-seq data (Table EXPRESSION / LABELING:
PHENOTYPE:
|
|
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |