- Title
-
Prdm8 regulates pMN progenitor specification for motor neuron and oligodendrocyte fates by modulating Shh signaling response
- Authors
- Scott, K., O'Rourke, R., Gillen, A., Appel, B.
- Source
- Full text @ Development

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
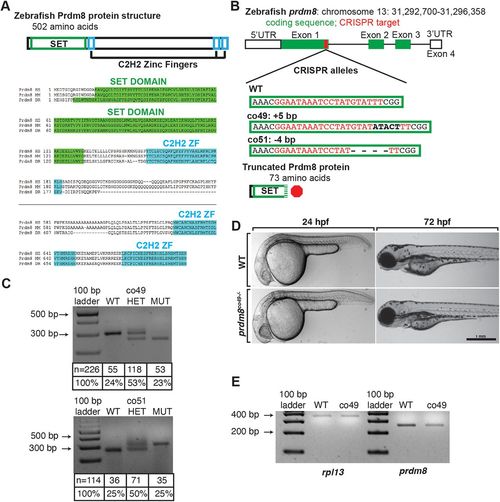
Generation and characterization of prdm8 loss-of-function mutations. (A) Zebrafish Prdm8 protein structure is depicted as an empty black box with the SET domain highlighted in green and the C2H2 zinc-finger domains in blue. Alignment of Prdm8 amino acid sequences from human (HS), mouse (MM), and zebrafish (DR). Conserved SET domain and C2H2 zinc-finger domains are shown as green or blue boxes, respectively. (B) Schematic representing prdm8 gene structure. The sequence targeted for CRISPR/Cas9-mediated mutagenesis is marked by a red line in exon 1. The wild-type sequence CRISPR target sequence is shown below in red text and the co49 insertion and co51 deletion are shown in bold or dashes, respectively. Both mutations are predicted to produce 73 amino acid proteins truncated at the C-terminal end of the SET domain. (C) Images of agarose gels showing prdm8 DNA fragmentation following dCAPS genotyping of homozygous wild-type (WT), heterozygous and homozygous mutant embryos with sample genotype frequencies. (D) Representative images of living 24 and 72 hpf wild-type and prdm8co49−/− embryos. (E) Image of RT-PCR gel, showing decreased expression of prdm8 mRNA in prdm8 mutant embryos compared with control, with no difference in expression of the control transcript rpl13. PHENOTYPE:
|
|
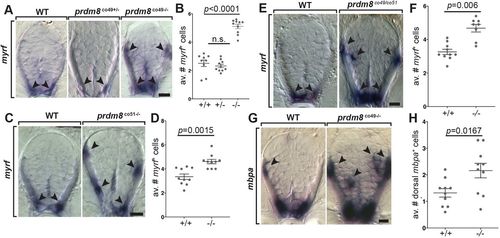
prdm8 mutant larvae have excess oligodendrocytes. (A,C,E,G) Representative trunk spinal cord transverse sections obtained from 72 hpf larvae showing mRNA expression patterns detected by ISH. Images and quantification of myrf expression in wild-type (WT), heterozygous and homozygous co49 mutant larvae (A,B), wild-type and homozygous co51 mutant larvae (C,D), and wild-type and co49/co51 mutant larvae (E,F). Arrowheads mark myrf+ oligodendrocytes. (G,H) Images of mbpa expression and quantification of dorsal mbpa+ oligodendrocytes in wild-type and homozygous co49 mutant larvae. Arrowheads denote mbpa+ oligodendrocytes. n=10 larvae for each genotype except wild type in B (n=9) and co49/co51 mutant larvae in F (n=8). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by Mann–Whitney U test (B,D,F) and an unpaired, two-tailed Student's t-test (H). n.s., not significant. Scale bars: 10 μm. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
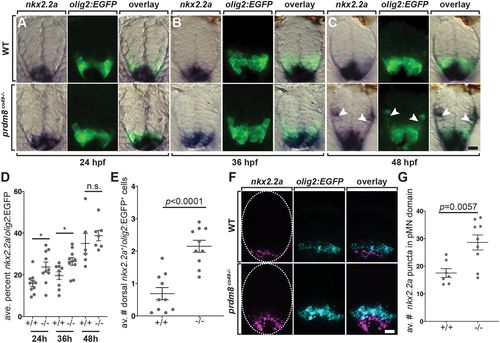
pMN cells prematurely express nkx2.2a in prdm8 mutant embryos. (A-C) Representative transverse sections of trunk spinal cord (dorsal up) showing prdm8 RNA (blue) and olig2:EGFP (green) expression. Developmental stages are noted at the bottom. Arrowheads indicate dorsally migrated oligodendrocyte lineage cells. (D) The area of nkx2.2a expression in the pMN domain is greater at 24 (n=10) and 36 hpf (co49, n=9; wild type, n=10) in prdm8 mutants embryos compared with controls and there is no difference at 48 hpf (co49 n=6; wild type, n=7). (E) prdm8co49−/− (n=10) have more dorsal OPCs (nkx2.2a+/olig2:EGFP+) than wild-type embryos (n=10) at 48 hpf. (F) Representative transverse trunk spinal cord sections obtained from 28 hpf embryos processed for fluorescent ISH to detect olig2 (blue) and nkx2.2a (pink) mRNA. Dashed ovals outline the spinal cord. (G) More nkx2.2a puncta are located within the olig2+ pMN domain of prdm8co49−/− embryos (n=9) compared with wild-type embryos (n=7). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test. n.s., not significant. *P<0.05. Scale bars: 10 μm. |
|
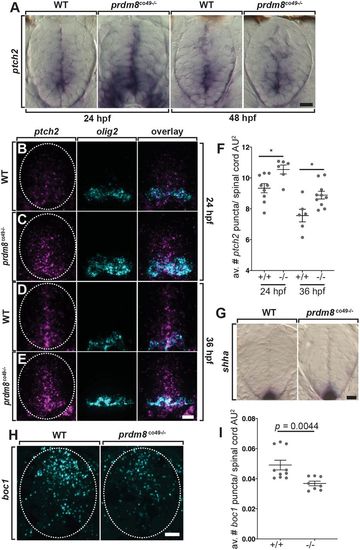
Spinal cord cells of prdm8 mutant embryos have elevated Shh signaling activity. (A) Representative transverse sections of trunk spinal cords obtained from 24 and 48 hpf wild-type (WT) and prdm8co49−/− embryos (dorsal up) showing ptch2 RNA expression. (B-E) Representative transverse trunk spinal cord sections processed for fluorescent ISH to detect olig2 (blue) and ptch2 (pink) mRNA at 24 hpf (B,C) and 36 hpf (D,E). (F) prdm8co49−/− embryos have more ptch2 puncta per AU2 of spinal cord at 24 hpf (n=6) and 36 hpf (n=10) than wild-type embryos at 24 hpf (n=9) and 36 hpf (n=6). (G) Representative transverse sections of trunk spinal cord (dorsal up) showing shha RNA expression in 24 hpf wild-type and prdm8co49−/− embryos. (H) Representative transverse trunk spinal cord sections processed for fluorescent ISH to detect boc1 (blue) mRNA at 24 hpf. (I) Wild-type embryos (n=6) have more boc1 puncta per AU2 of spinal cord at 24 hpf than prdm8co49−/− embryos (n=8). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test (F) and by Mann-Whitney U test (I). *P<0.001. Dashed oval outlines the spinal cord boundary. Scale bars: 10 μm. |
|
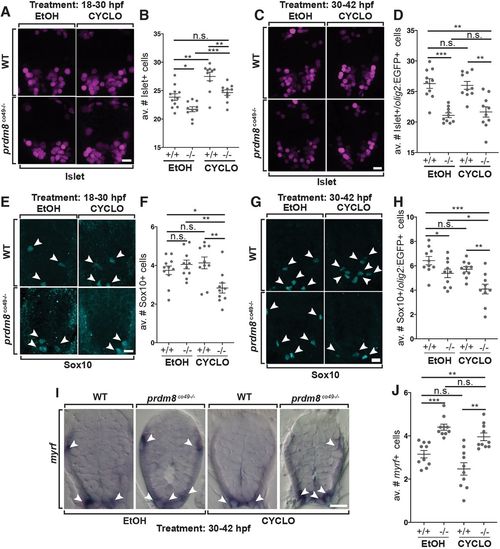
Shh inhibition rescues the motor neuron but not oligodendrocyte phenotypes of prdm8 mutant embryos. (A,C,E,G) Representative images of trunk spinal cord sections from 48 hpf embryos treated with 0.5 μM cyclopamine (CYCLO) or ethanol (EtOH) from 18 to 30 hpf (A,E) or 30 to 42 hpf (C,G), and processed to detect Isl (A,C) or Sox10 (E,G) expression. (A,B) Wild-type (WT) embryos treated with EtOH control solution and prdm8 mutant embryos treated with cyclopamine have similar numbers of motor neurons. (C,D) There are fewer motor neurons (Islet+) in prdm8co49−/− embryos treated with EtOH and cyclopamine compared with wild-type embryos treated with EtOH. (E,F) There are fewer OPCs (Sox10+; arrowheads) in prdm8co49−/− embryos treated with cyclopamine and no difference in OPCs prdm8co49−/− embryos treated with EtOH compared with wild-type embryos treated with EtOH. (G,H) There are fewer OPCs (Sox10+; arrowheads) in prdm8co49−/− embryos treated with cyclopamine and slightly fewer OPCs in prdm8co49−/− embryos treated with EtOH compared with wild-type embryos treated with EtOH. (I) Representative trunk spinal cord transverse sections obtained from 72 hpf larvae treated with 0.5 μM cyclopamine or EtOH from 30 to 42 hpf showing myrf mRNA expression detected by in situ RNA hybridization. (I,J) prdm8co49−/− embryos treated with EtOH or cyclopamine have more oligodendrocytes (myrf+; arrowheads) than wild-type embryos treated with EtOH. n=10 for all genotypes and treatments except for wild-type embryos treated with EtOH (n=11) (A,E). Data are mean±s.e.m. with individual data points indicated. Statistical significance was evaluated by an unpaired, two-tailed Student's t-test. *P<0.05; **P<0.001; ***P<0.0001; n.s., not significant. Scale bars: 10 μm. EXPRESSION / LABELING:
PHENOTYPE:
|