- Title
-
Identification of slit3 as a locus affecting nicotine preference in zebrafish and human smoking behaviour
- Authors
- García-González, J., Brock, A.J., Parker, M.O., Riley, R.J., Joliffe, D., Sudwarts, A., Teh, M.T., Busch-Nentwich, E.M., Stemple, D.L., Martineau, A.R., Kaprio, J., Palviainen, T., Kuan, V., Walton, R.T., Brennan, C.H.
- Source
- Full text @ Elife
|
( PHENOTYPE:
|
|
( |
|
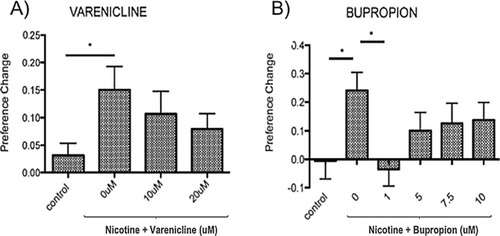
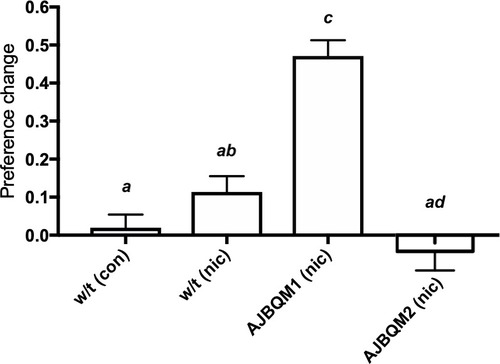
AJBQM1 and AJBQM2 siblings, not included in the screen (n = 10 for AJBQM1; n = 14 for AJBQM2), AJBQM1 significantly differed from the parental strain, Tupfel longfin (TLF) wild type (w/t) saline control (n = 17) and wild type nicotine exposed fish (n = 7). AJBQM2 differed from wild type nicotine exposed fish but not wild type saline controls. Different superscript letters indicate significant difference between groups (p<0.05), same superscript letters indicate no significant differences between groups. Bars indicate Mean + SEM. |
|
( PHENOTYPE:
|
|
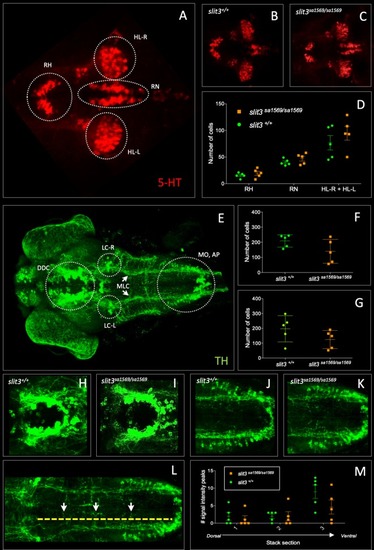
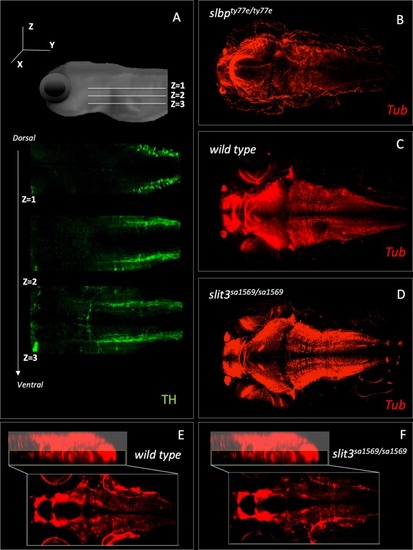
(A–D) Anti-5-HT, (E–M) anti-TH: (A) 5-HT-labelled neurons in wild type zebrafish brain. Circles indicate regions used for quantification of cell number in rostral hypothalamus (RH), inferior hypothalamic lobes (HL-R, HL-L) and raphe nucleus (RN). (B) Anti-5-HT labelled cells in slit3 wild type brain, (C) Anti-5-HT labelled cells in slit3sa1569 homozygous mutant brain. (D) Quantification of anti-5HT labelled cell number in wild type and slit3 mutant brains No significant differences were observed between wild type and slit3 mutant larvae. (E) Unprocessed maximum intensity projection of anti-TH-labelled whole mounted wild type zebrafish brain. Circles indicate areas used for quantification, or in the case of LC-R and LC-L, landmarks used as reference to determine the extension of the medial longitudinal catecholaminergic tract (MLC) used when quantifying the number of anti-TH labelled projections to the midline (panels L, M). (F) Cell quantification for diencephalic dopaminergic cluster (DDC). No significant differences were observed between wild type and slit3 mutant larvae. (G) Cell quantification for medulla oblongata interfascicular zone and vagal area, and area postrema (MO, AP). No significant differences were observed between wild type and slit3 mutant larvae. (H–K) Anti-TH labelled wild types and slit3sa1569. Zoomed-in visualization of diencephalic dopaminergic cluster (H–I) and medulla oblongata interfascicular zone and vagal area (J–K). (L–M) Quantification of catecholaminergic projections projecting to the midline. Examples of projections are indicated with yellow arrows. Projections were assessed from posterior to anterior using the locus coerulus and posterior extent of the raphe nucleus as landmarks (Panel L, yellow line) and from dorsal to ventral (Panel M, stacks 1–3). Figure 5—figure supplement 1 shows individual planes. n = 5 samples per genotype group. EXPRESSION / LABELING:
|
|
( |
|
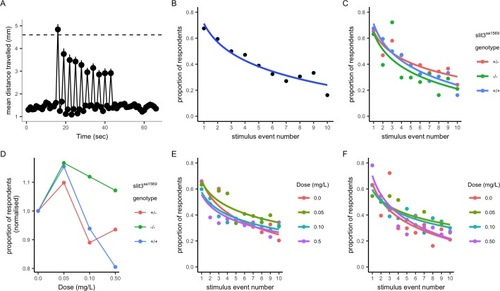
(A–B) Response and habituation to 10 stimuli with two seconds interval between stimuli in wild type, drug free zebrafish. Mean distances travelled were measured in one second time bins. Line indicates 4.6 mm, which corresponds to mean basal distance moved per second plus 2 standard deviations of the mean and was used to define respondents. The percentage of fish responding to the stimuli decreases with stimulus/tap number (Main effects of tap number p<0.05) 68% respond to the first tap; 16% respond to the last tap. Respondents are defined as fish moving more than 4.6 mm. (C) Proportion of responders across the ten stimuli in drug free individuals from each genotype: there was no significant effect of genotype on response across taps (p=0.34) or responsiveness (p=0.35) in drug free fish. (D) Mean percentage of responders across the ten stimuli (± SEM). Data are stratified by slit3sa1569 genotype and amisulpride dose normalised to response in absence of drug. The effect of amisulpride on habituation varies by genotype. (E, F) Proportion of individuals responding in each amisulpride dose condition in wild type and homozygous mutant fish, respectively. The interaction between amisulpride dose and stimulus event number had a significant effect on the proportion of responsive individuals in wild type individuals (p<0.05) but not homozygous mutants (p=0.16). PHENOTYPE:
|
|
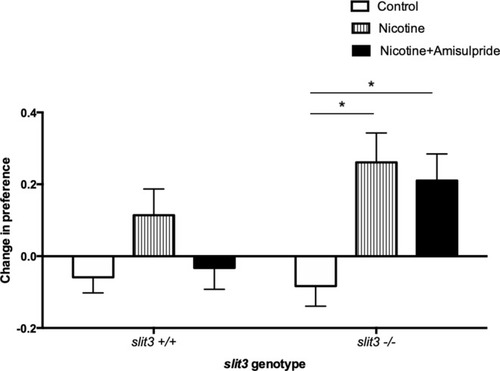
( |
|
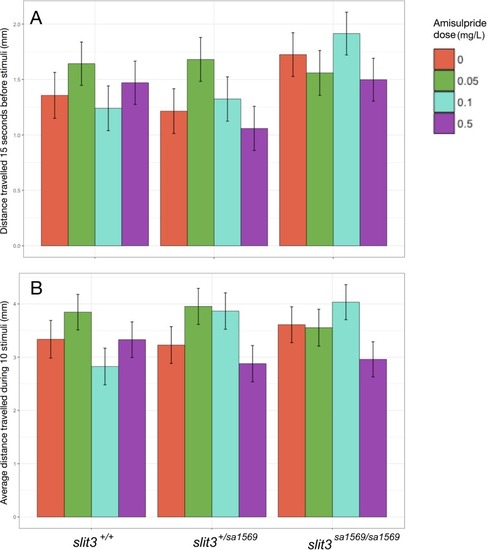
Bars represent mean (+ SEM). (n = 11–14 fish per group). *Two-way ANOVA followed by post-hoc Tukey tests (p<0.05). PHENOTYPE:
|
|
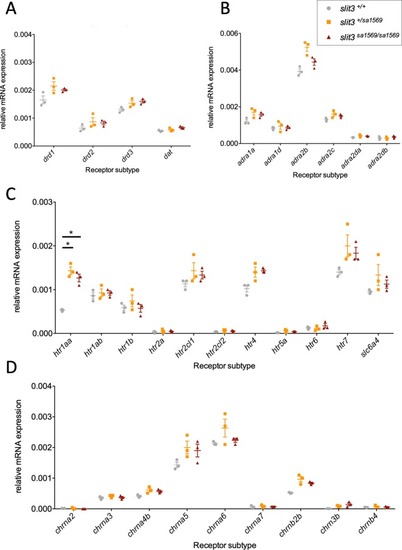
Quantitative PCR analysis of gene expression was performed for members of ( |
|
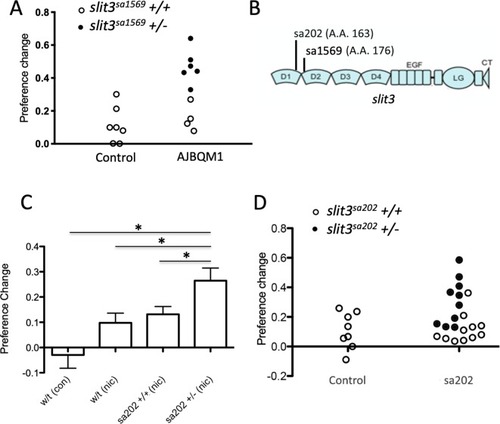
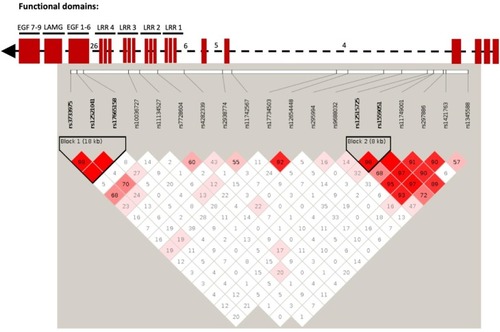
Numbers within each square indicate D’ values (white: D’<1, LOD <2; blue: D’=1, LOD <2; pink: D’<1, LOD ≥2; and bright red: D’=1, LOD ≥2). Top part of the figure shows domain organization of the SLIT protein based on the UCSC Genome Browser ( |
|
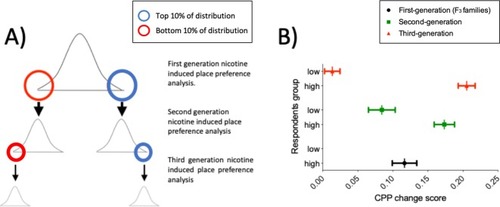
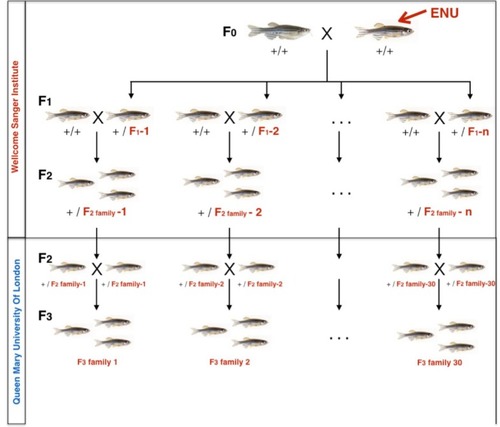
F2 ENU-mutagenized zebrafish, heterozygous for multiple mutations across the entire genome were obtained from the Wellcome Sanger Institute as part of the Zebrafish Mutation Project. At QMUL, heterozygous F2 fish were incrossed to generate 30 F3 families, each containing 10–20 nonsense or essential splice site mutations and about 1500 additional exonic and intronic point mutations. F3 Families were arbitrarily numbered 1–30. Expected mutation rate and type of mutations in coding regions are specified on the right hand side. |