- Title
-
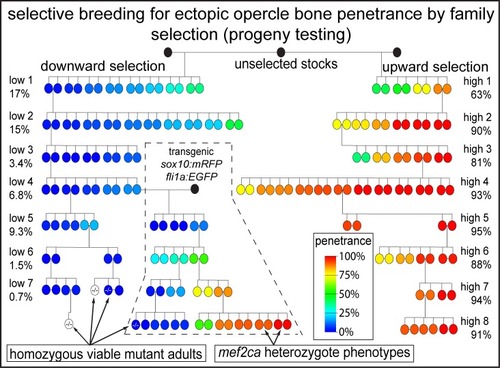
Selective breeding modifies mef2ca mutant incomplete penetrance by tuning the opposing Notch pathway
- Authors
- Sucharov, J., Ray, K., Brooks, E.P., Nichols, J.T.
- Source
- Full text @ PLoS Genet.
|
Full-sibling heterozygous |
|
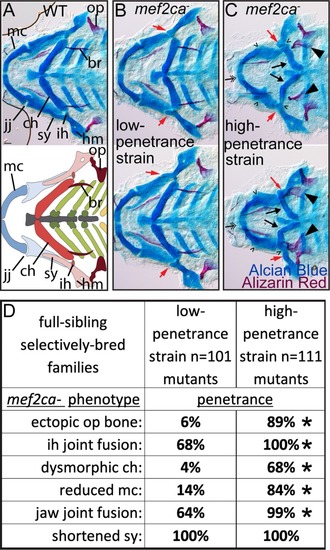
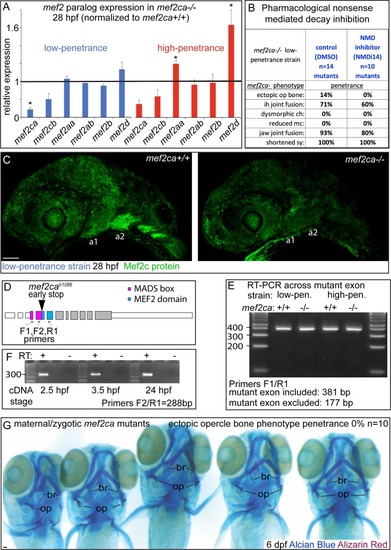
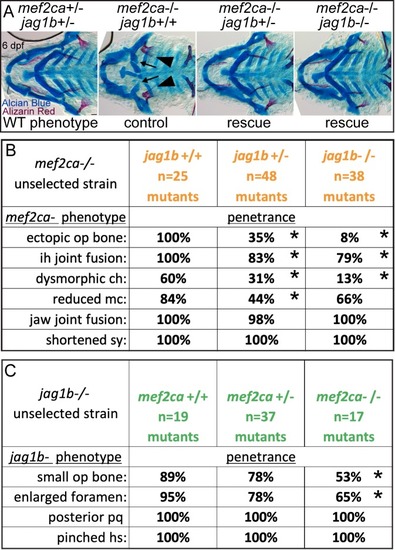
(A) Wild-type 6 days post fertilization (dpf) zebrafish larvae were stained with Alcian Blue and Alizarin Red to label cartilage and bone then flat mounted and imaged. The following craniofacial skeletal elements are indicated in the micrograph and graphic: opercle bone (op), branchiostegal ray (br), Meckel’s (mc), ceratohyal (ch), symplectic (sy) and hyomandibular (hm) cartilages, interhyal (ih) and jaw (jj) joints. Scale bar: 50 μm. (B, C) Penetrance of most phenotypes associated with PHENOTYPE:
|
|
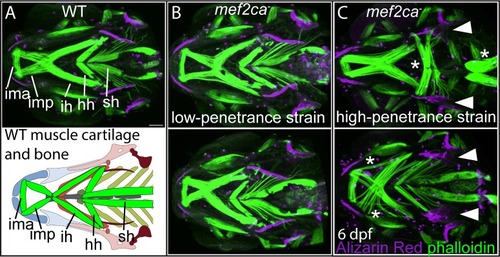
6 dpf larvae were stained with Alizarin Red (magenta) and fluorescently labelled phalloidin (green) to mark bones and muscles, respectively. (A) The following craniofacial muscles are indicated in the micrograph and graphic: intermandibularis ant. (ima), intermandibularis post. (imp), interhyal (ih), hyohyal (hh), sternohyoideus (sh). Scale bar: 50 μm. (B) In the low-penetrance strain, PHENOTYPE:
|
|
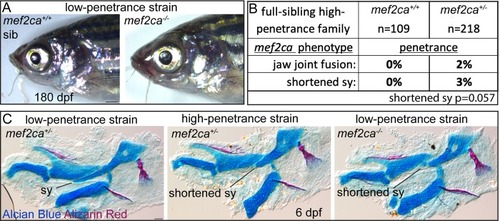
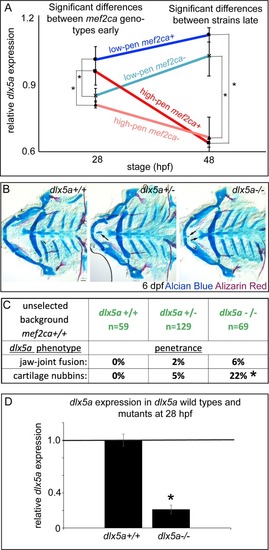
(A) Low-penetrance PHENOTYPE:
|
|
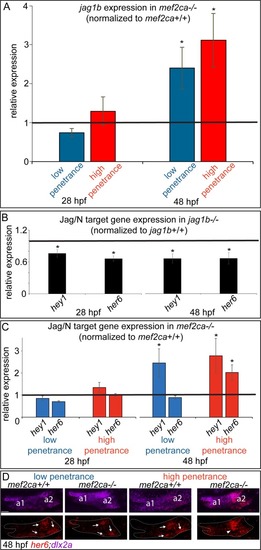
(A) Head expression of all |
|
(A) Head expression of |
|
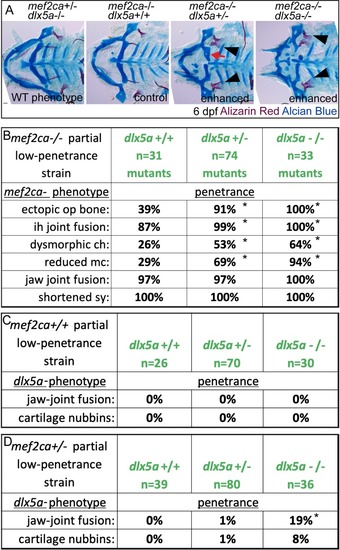
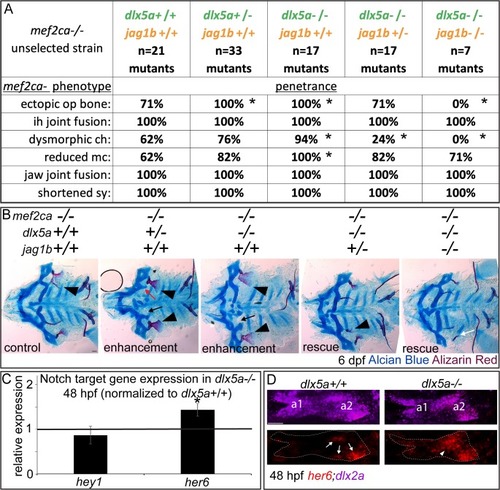
(A) Animals double heterozygous for |
|
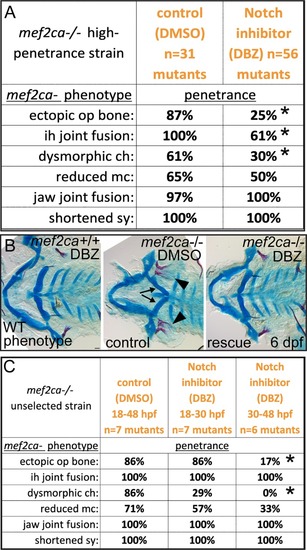
(A) Animals heterozygous for PHENOTYPE:
|
|
(A) Animals double heterozygous for |
|
(A) Head expression of |
|
(A) Animals triple heterozygous for |
|
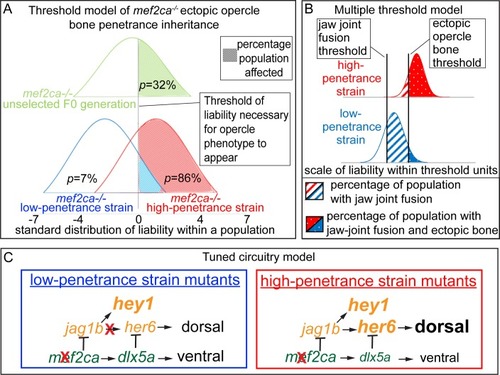
(A) The liability-threshold model illustrates the inheritance of ectopic bone penetrance in |