- Title
-
AP endonuclease 1 (Apex1) influences brain development linking oxidative stress and DNA repair
- Authors
- Pei, D.S., Jia, P.P., Luo, J.J., Liu, W., Strauss, P.R.
- Source
- Full text @ Cell Death Dis.
|
|
|
PHENOTYPE:
|
|
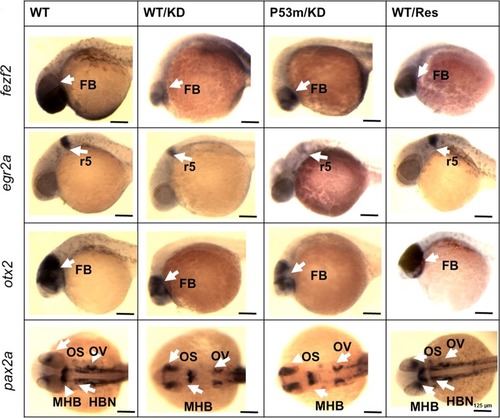
Whole mount in situ hybridization shows aberrant distribution of critical brain markers after Apex1 knockdown. Rescue was achieved by co-injection of transcript for human |
|
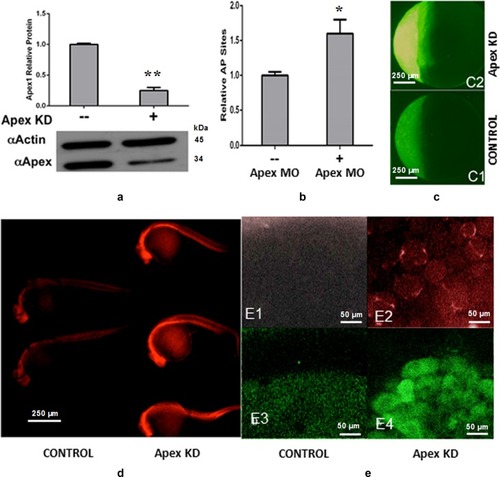
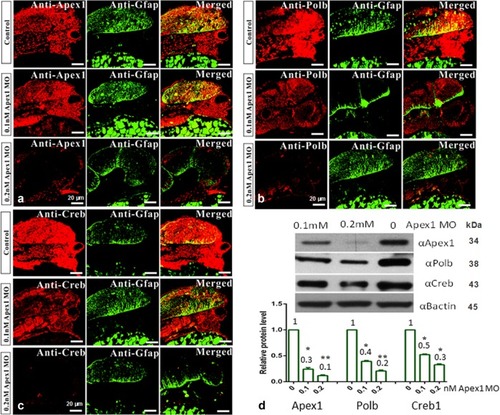
Standard fluorescent immunohistochemical staining was performed, and recorded by confocal microscopy (×40 magnification). Zebrafish embryos at the 1-cell stage were injected with control MO, 0.1, or 0.2 mM Apex1 MO, and allowed to develop for an additional 48 h before they were fixed and processed as described in Methods. Green fluorescence represents the expression pattern for glial fibrillary acidic protein (GFAP), while red fluorescence indicates the expression pattern for the indicated protein: Apex1 ( |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|