- Title
-
nkx genes establish SHF cardiomyocyte progenitors at the arterial pole and pattern the venous pole through Isl1 repression
- Authors
- Colombo, S., de Sena-Tomás, C., George, V., Werdich, A.A., Kapur, S., MacRae, C.A., Targoff, K.L.
- Source
- Full text @ Development
|
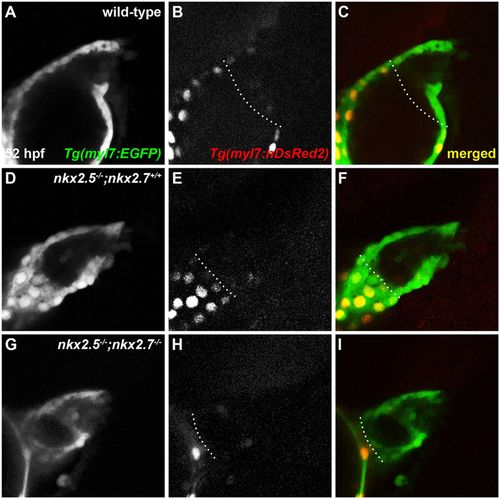
OFT size and CM numbers are reduced in nkx mutants. (A-I) Lateral view, anterior towards the right, at 52 hpf. Representative reconstructions of four z-stacks from a developmental timing assay show diminished cell numbers in the OFT of wild-type (A-C), nkx2.5−/−;nkx2.7+/+ (D-F) and nkx2.5−/−;nkx2.7−/− (G-I) embryos carrying Tg(myl7:EGFP) and Tg(-5.1myl7:nDsRed2). |
|
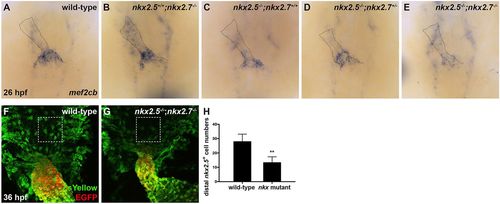
Nkx genes delimit the OFT progenitor pool. (A-E) Dorsal view, anterior towards the top, at 26 hpf. In situ hybridization depicts diminished expression of mef2cb, which labels aSHF-derived CM progenitors, in representative nkx2.5−/−;nkx2.7+/+ (n=6) (C), nkx2.5−/−;nkx2.7+/− (n=3) (D) and nkx2.5−/−;nkx2.7−/− (n=5) (E) embryos compared with wild-type (n=15) (A) and nkx2.5+/+;nkx2.7−/− (n=5) (B) embryos. (F,G) Ventral view of the arterial pole region at 36 hpf. Confocal projections of immunohistochemistry for ZsYellow (green) and EGFP (red) in embryos carrying Tg(nkx2.5:ZsYellow) and Tg(myl7:EGFP) illustrate decreased contribution of nkx2.5+ aSHF-derived CM progenitors (ZsYellow+ EGFP−) to the arterial pole in nkx2.5−/−;nkx2.7−/− (G) compared with wild-type (F) embryos (boxed areas). (H) Quantification of ZsYellow+ EGFP− cells at the arterial pole in wild-type (n=9) and nkx mutant (n=8) embryos reveals a statistically significant decrease in this population following the loss of Nkx gene function. Student's t-test was used to determine statistical significance. Mean and s.e.m. of each data set are shown (**P<0.0001). |
|
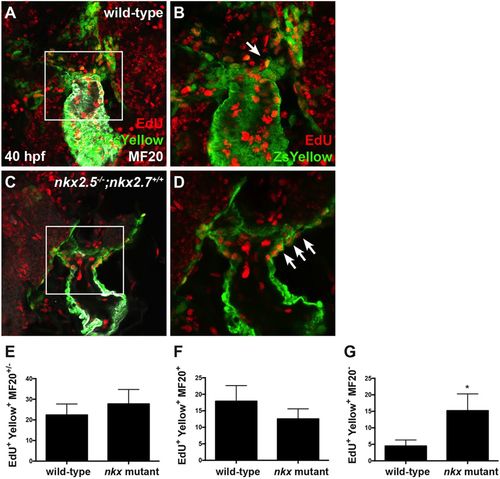
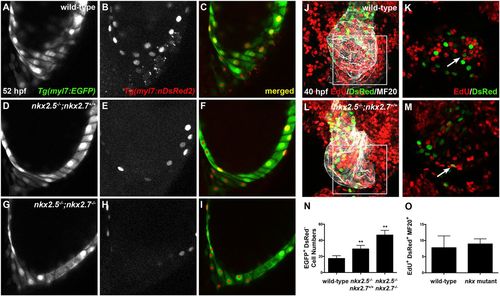
Nkx genes regulate OFT progenitor proliferation timing and differentiation. (A-D) Ventral view of the arterial pole region at 40 hpf. Confocal projections of immunohistochemistry for EdU (red), ZsYellow (green) and MF20 (gray) in embryos carrying Tg(nkx2.5:ZsYellow) following EdU incubation at 18 hpf in wild-type (A,B) and nkx2.5−/−;nkx2.7+/+ (C,D) embryos. White arrows indicate EdU+ ZsYellow+ cells. The boxed areas in A,C are shown at higher magnification in B,D. (E-G) Proliferation indices showing a statistically significant increase in undifferentiated, ZsYellow+ MF20− progenitors in nkx mutant (n=6) compared with wild-type (n=6) embryos (G); the proliferation index is comparable in the proximal, ZsYellow+ MF20+ CM population contributing to the arterial pole (F) as well as in the total OFT, ZsYellow+ MF20+/− CM population (E). Student's t-test was used to determine statistical significance. The mean and s.e.m. of each data set are shown (*P<0.005). |
|
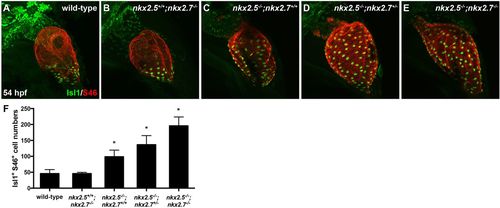
Nkx genes inhibit Isl1 expression at the sinus venosus. (A-E) Lateral view, anterior towards the top, at 54 hpf in wild-type (A), nkx2.5+/+;nkx2.7−/− (B), nkx2.5−/−;nkx2.7+/+ (C), nkx2.5−/−;nkx2.7+/− (D) and nkx2.5−/−;nkx2.7−/− (E) embryos. Representative confocal images of immunofluorescence for Isl1 (green) and S46 (red) demonstrate that Isl1 is restricted to the venous pole of wild-type (A) and nkx2.5+/+;nkx2.7−/− (B) embryos. However, Isl1 expression expands throughout the atrium in nkx mutants (C-E). (F) Quantification of Isl1+ S46+ CMs is depicted showing a gradual increase in cell number with a progressive loss of nkx alleles. Mean and s.e.m. of each data set are shown (*P<0.005) in wild-type (n=16), nkx2.5+/+;nkx2.7−/− (n=2), nkx2.5−/−;nkx2.7+/+ (n=6), nkx2.5−/−;nkx2.7+/− (n=10) and nkx2.5−/−;nkx2.7−/− (n=4) embryos. Student's t-test was used to determine statistical significance. |
|
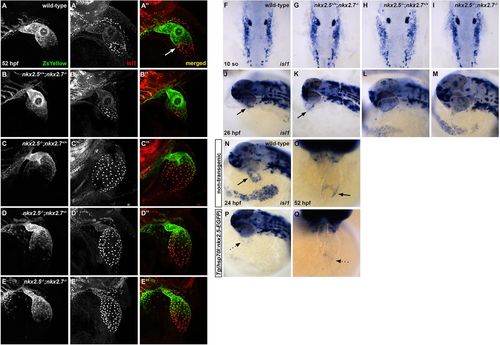
Nkx2.5 is not required for isl1+ progenitor specification, but is sufficient to repress isl1 at the venous pole. (A-E″) Representative confocal images, lateral view, anterior towards the top, at 52 hpf. Immunostaining for Isl1 (red) in embryos carrying Tg(nkx2.5:ZsYellow) demonstrates that Isl1 is restricted to the venous pole and Nkx2.5 expression is absent in this region in wild-type (white arrow) (A-A″) and nkx2.5+/+;nkx2.7−/− (B-B″) embryos. However, Isl1 expression is derepressed in the atrium of nkx2.5−/−;nkx2.7+/+ (C-C″) and nkx2.5−/−;nkx2.7+/− (D-D″) embryos, and expands towards the outflow tract in nkx2.5−/−;nkx2.7−/− (E-E″) embryos. (F-I) Dorsal view, anterior towards the top, at 10 somites. In situ hybridization shows no variation in ALPM isl1 expression between wild-type (F) and nkx mutant (G-I) embryos. (J-M) Lateral view, anterior towards the left, at 26 hpf. In situ hybridization demonstrates that isl1 expression is restricted to a ring at the venous pole of the heart tube in wild-type (J) and nkx2.5+/+;nkx2.7−/− (K) embryos. However, isl1 expression expands to the atrium in nkx2.5−/−;nkx2.7+/+ embryos (L) and throughout the entire heart tube in nkx2.5−/−;nkx2.7−/− embryos (M). Black arrows indicate the ring of isl1 expression near the IFT (J,K). (N-Q) Lateral view, anterior towards the left, at 24 hpf (N,P) and ventral view, anterior towards the top, at 52 hpf (O,Q) in non-transgenic (N,O) and Tg(hsp70l:nkx2.5-EGFP) (P,Q) embryos. In situ hybridization demonstrates that isl1 expression is restricted to a ring at the venous pole (black arrows) at 24 hpf (n=9) (N) and 52 hpf (n=7) (O), and that overexpression of nkx2.5 inhibits isl1 expression specifically in this region (dotted arrows) at 24 hpf (n=9/11) (P) and 52 hpf (n=8/9) (Q). EXPRESSION / LABELING:
|
|
Nkx genes regulate cardiomyocyte accretion to the atrium. (A-I) Lateral images, anterior towards the top, at 52 hpf. Representative reconstructions of 10 z-stacks from a developmental timing assay highlight the venous pole of wild-type (A-C), nkx2.5−/−;nkx2.7+/+ (D-F) and nkx2.5−/−;nkx2.7−/− (G-I) embryos carrying Tg(myl7:EGFP) (green) and Tg(-5.1myl7:nDsRed2) (red). Asterisks indicate EGFP+ DsRed− cells. (J-M) Ventral view, anterior towards the top, at 40 hpf of wild-type (J,K) and nkx2.5−/−;nkx2.7+/+ (L,M) embryos carrying Tg(-5.1myl7:nDsRed2). Confocal projections (J,L) of immunohistochemistry for EdU (red), DsRed (green) and MF20 (gray) following EdU incubation at 18 hpf and four z-stack reconstructions (K,M) were used to count EdU+ DsRed+ MF20+ CMs in the atrium. The boxed areas in J,L are shown at higher magnification in K,M, respectively. White arrows indicate EdU+ DsRed+ CMs. (N,O) Quantification of EGFP+ DsRed− cells at the venous pole for wild-type (n=12), nkx2.5−/−;nkx2.7+/+ (n=5) and nkx2.5−/−;nkx2.7−/− (n=4) embryos, and proliferation indices in wild-type (n=5) and nkx mutant (n=3) embryos. Student's t-test was used to determine statistical significance. Mean and s.e.m. of each data set are shown (**P<0.0001). There is no statistically significant difference between proliferation indices in the DsRed+ MF20+ populations of wild-type and nkx mutant embryos. |
|
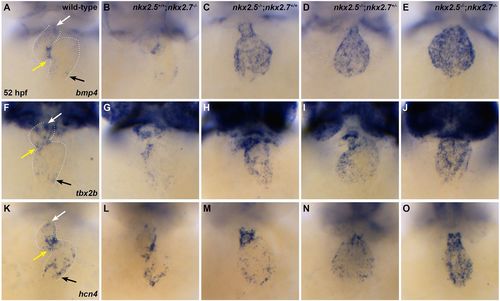
Nkx genes regulate SAN markers. (A-O) Ventral view, anterior towards the top, at 52 hpf. Dotted white lines illustrate the cardiac silhouettes. In situ hybridization for bmp4 (A-E), tbx2b (F-J) and hcn4 (K-O). Expression of bmp4 is restricted to the OFT (white arrow), AVC (yellow arrow) and IFT (black arrow) in wild-type (n=7/7) (A) and nkx2.5+/+;nkx2.7−/− (n=4/4) (B) embryos. However, its expression is expanded throughout the cardiac chambers in nkx2.5−/−;nkx2.7+/+ (n=3/4) (C), nkx2.5−/−;nkx2.7+/− (n=3/3) (D) and nkx2.5−/−; nkx2.7−/− (n=3/3) (E) embryos. Similar findings are observed in the wild-type (n=6/7) (F), nkx2.5+/+;nkx2.7−/− (n=3/4) (G), nkx2.5−/−;nkx2.7+/+ (n=3/3) (H), nkx2.5−/−;nkx2.7+/− (n=2/2) (I) and nkx2.5−/−;nkx2.7−/− (n=2/2) (J) embryos using the tbx2b probe, and in wild-type (n=8/8) (K), nkx2.5+/+;nkx2.7−/− (n=5/6) (L), nkx2.5−/−;nkx2.7+/+ (n=9/11) (M), nkx2.5−/−;nkx2.7+/− (n=3/3) (N) and nkx2.5−/−;nkx2.7−/− (n=3/3) (O) embryos using the hcn4 probe. |
|
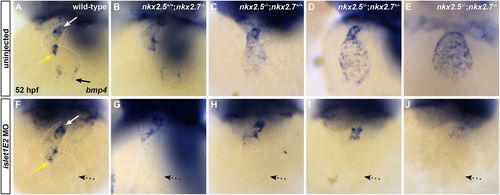
isl1 acts downstream of Nkx genes at the venous pole. (A-J) Ventral view, anterior towards the top, at 52 hpf. Dotted white lines illustrate the cardiac silhouettes. In situ hybridization for bmp4 (A-J) in wild-type (A,F), nkx2.5+/+;nkx2.7−/− (B,G), nkx2.5−/−;nkx2.7+/+ (C,H), nkx2.5−/−;nkx2.7+/− (D,I) and nkx2.5−/−;nkx2.7−/− (E,J) embryos. Inhibition of isl1 expression using islet1E2 MO in wild-type (n=4/4) (compare A with F) and nkx2.5+/+;nkx2.7−/− (n=4/4) (compare B and G) embryos leads to downregulation of bmp4 expression specifically at the venous pole of the heart (black arrow) without affecting the AVC (yellow arrow) and OFT (white arrow) expression. In nkx2.5−/−;nkx2.7+/+ (n=2/2), nkx2.5−/−;nkx2.7+/− (n=5/6) and nkx2.5−/−;nkx2.7−/− (n=3/3) embryos, isl1 inhibition rescues the misregulation of bmp4 expression in the atrium (dotted arrows), yet AVC and OFT expression patterns remain unchanged (compare C-E with H-J). |
|
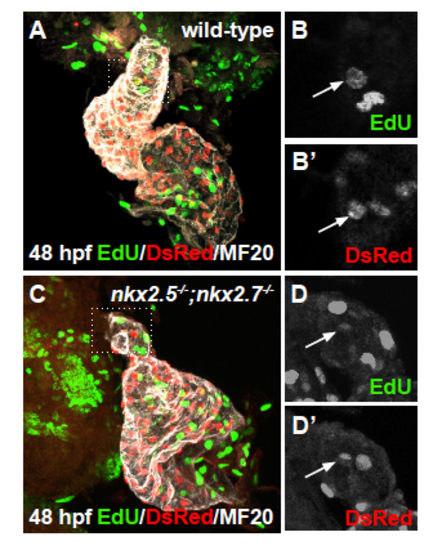
Proliferation of differentiated arterial pole CMs is not affected (A-D) Ventral view, anterior to the top, of the arterial pole region at 48 hpf. Confocal projections of immunohistochemistry for EdU (green), DsRed (red), and MF20 (gray) in wild-type (A,B-B’) and nkx2.5-/-;nkx2.7-/- (C,D-D’) embryos carrying Tg(- 5.1myl7:nDsRed2) following EdU incubation at 24 hpf. Representative images highlight the detection of proliferating nuclei in the OFT myocardium. EXPRESSION / LABELING:
|
|
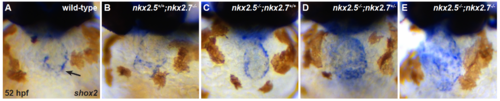
nkx genes regulate shox2 (A-E) Ventral view, anterior to the top, at 52 hpf illustrates in situ hybridization for shox2. Expression of shox2 is restricted to the IFT in wild-type (black arrow) (n = 11/13) (A) and nkx2.5+/+;nkx2.7-/- (n = 5/6) (B) embryos. However, its expression is expanded throughout the cardiac chambers in nkx2.5-/-;nkx2.7+/+ (n = 9/10) (C), nkx2.5- /-;nkx2.7+/- (n = 12/13) (D), and nkx2.5-/-;nkx2.7-/- (n = 5/7) (E) embryos. |