- Title
-
hnf1b genes in zebrafish hindbrain development
- Authors
- Choe, S.K., Hirsch, N., Zhang, X., and Sagerström, C.G.
- Source
- Full text @ Zebrafish
|
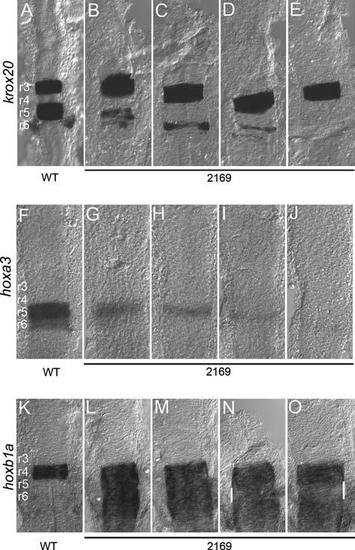
Variable gene expression in the hindbrain of hnf1bhi2169 mutant embryos. Wild-type (A, F, K) and hnf1bhi2169 (B–E, G–J, L–O) embryos were assayed for expression of krox20 (A–E), hoxa3 (F–J), and hoxb1a (K–O) by in situ hybridization. All panels are dorsal views of flat-mounted hindbrains with anterior to the top. Rhombomere numbering is indicated in panels (A), (F), and (K). White bars in panels (N) and (O) indicate regions of reduced hoxb1a expression. EXPRESSION / LABELING:
PHENOTYPE:
|
|
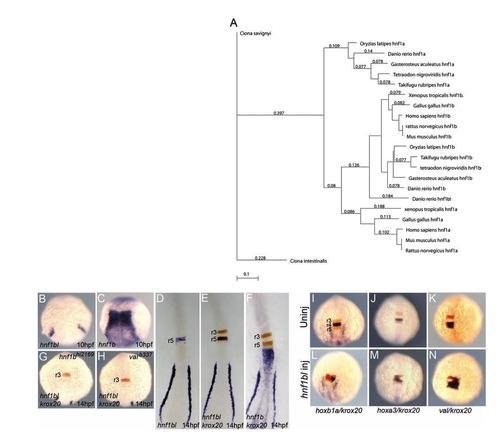
A novel hnf1bl gene expressed in r5. (A) Phylogenetic tree demonstrating that hnf1bl clusters with hnf1b genes rather than hnf1a genes. (B–H) Wild-type (B–F), hnf1bhi2169 (G), and valb337 (H) embryos were assayed for expression of hnf1bl (B, D), hnf1b (C), hnf1bl+krox20 (E, G, H), or hnf1b+krox20 (F) by in situ hybridization. Note that hnf1b and hnf1bl are detected in blue, while krox20 is detected in red. (B), (C), (G), and (H) are whole mounts while (D), (E), and (F) are flat mounts. Anterior is to the top in all panels. (I–N) Uninjected (I–K) and hnf1bl-injected (L–N) wild-type embryos were assayed for expression of hoxb1a+krox20 (I, L), hoxa3+krox20 (J, M), or val+krox20 (K, N). Note that hoxb1a, hoxa3, and val are detected in blue, while krox20 is detected in red. All embryos are whole mounts in dorsal view with anterior to the top. EXPRESSION / LABELING:
PHENOTYPE:
|
|
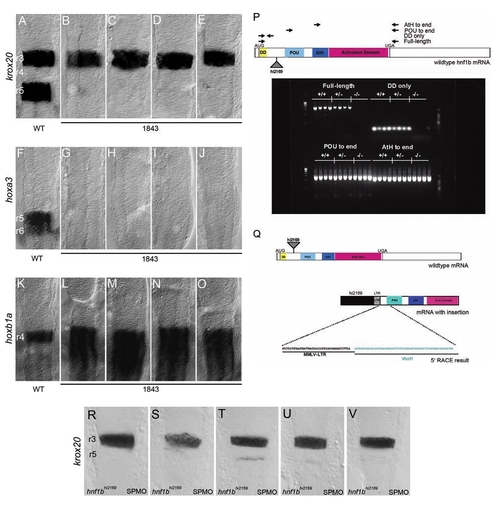
(A–O) Wild-type (A, F, K) and hnf1b1843 (B–E, G–J, L–O) embryos were assayed for expression of krox20 (A–E), hoxa3 (F–J), and hoxb1a (K–O) by in situ hybridization. All panels are dorsal views of flat-mounted hindbrains with anterior to the top. Rhombomere numbering is indicated in panels (A), (F), and (K). (P) Wild-type (+/+), heterozygous (+/-), and homozygous hnf1bhi2169 mutant (-/-) embryos were assayed for presence of hnf1b transcripts by RT-PCR. Virus integration site and locations of primers used for RT-PCR analysis are indicated in diagram at top. RT-PCR products were resolved by gel electrophoresis, and the resulting gel is shown at the bottom. (Q) Primer extension analysis of virus-hnf1b transcript. Top panel shows structure of wild-type hnf1b mRNA and indicates retroviral insertion site (triangle) in the hnf1bhi2169 allele. Bottom panel shows putative viral-mRNA fusion transcript and indicates sequence obtained from primer extension analysis. (R–V) hnf1bhi2169 embryos injected with an hnf1b spliceblocking morpholino (SPMO) show near-complete loss of krox20 expression in r5 compared to Figure 1B–E. EXPRESSION / LABELING:
PHENOTYPE:
|
|
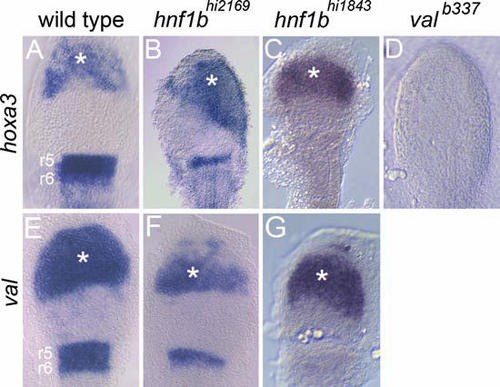
hoxb1b activates r5/r6 gene expression independently of hnf1b in the anterior embryo, but not in the hindbrain. Wild-type (A, E), hnf1bhi2169 (B, F), hnf1bhi1843 (C, G), and valb337 (D) embryos were injected with hoxb1b and meis3 mRNA and assayed for expression of hoxa3 (A–D) and val (E–G). Ectopic gene expression anterior to the hindbrain is indicated by asterisks. All panels are flat-mounted dorsal views of the anterior embryo with anterior to the top. |
|
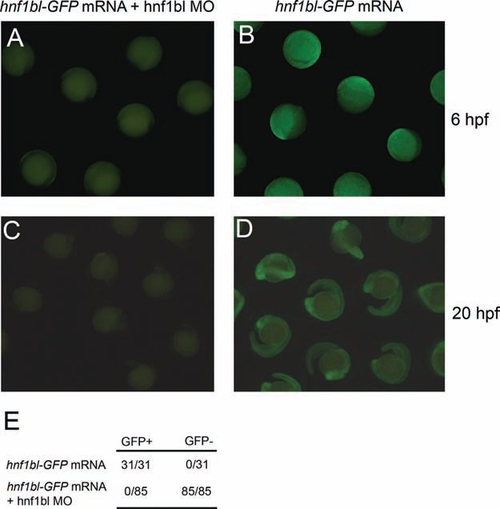
(A–D) Wild-type zebrafish embryos were injected with 100 pg mRNA encoding an Hnf1bl-GFP fusion protein either alone (B, D), or together with 1mM hnf1bl MO (A, C). Embryos were scored for GFP expression at 6 hpf (A, B) or 20 hpf (C, D). (E) Data from a representative experiment showing that 31/31 embryos injected with hnf1bl-GFP mRNA are GFP+, and 0/85 embryos injected with hnf1bl-GFP mRNA together with hnf1bl MO are GFP+. |