Fig. S4
- ID
- ZDB-FIG-190604-59
- Publication
- Lan et al., 2019 - TETs Regulate Proepicardial Cell Migration through Extracellular Matrix Organization during Zebrafish Cardiogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
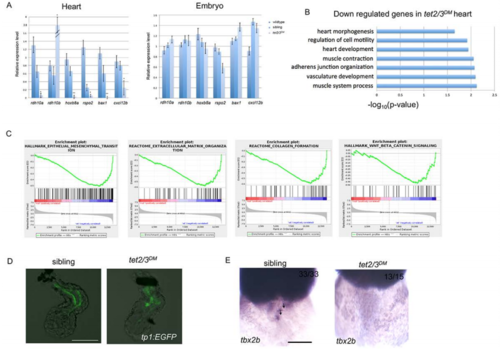
tet2/3DM Larvae Show Cardiac AVC Defects at 48-hpf. Related to Figure 3 and Figure 4. (C) Gene set enrichment analysis shows down-regulated biological pathways in 48-hpf tet2/3DM hearts compared to wildtype hearts by RNA sequencing using isolated hearts. |