Fig. S1
- ID
- ZDB-FIG-071204-34
- Publication
- Li et al., 2007 - Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos
- Other Figures
- All Figure Page
- Back to All Figure Page
|
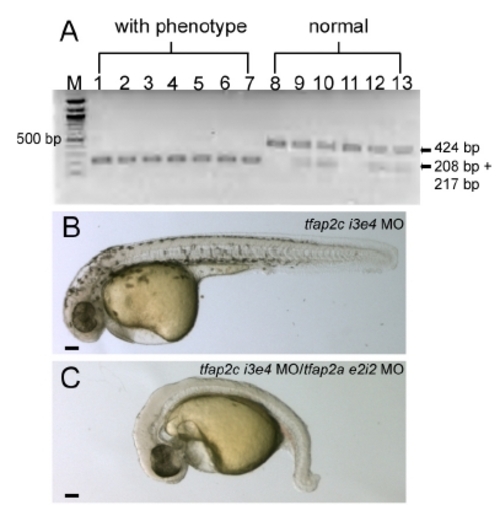
A PCR-based genotype assay of tfap2c MO-injected tfap2a mutant embryos. The low mutation introduces a Blp1 restriction site into exon 5 of tfap2a cDNA (Knight et al., 2003). tfap2c e3i3 MO was injected into 31 embryos derived from tfap2a heterozygous mutant parents. PCR on genomic DNA harvested from individual embryos revealed that all embryos with the strong phenotype harbored two copies of the Blp1-sensitive allele of tfap2a, while embryos with normal phenotype harbored 1 or 0 copies of this allele. All 31 embryos were assayed; this gel shows results from all 7 embryos with the strong phenotype (lanes 1–7) and 6 embryos with normal morphology. M, 100 bp ladder. (B, C) Lateral views of live embryos at 30 hpf. In B, a wild-type embryo injected with tfap2c i3e4 MO (hereafter, control embryo) with normal morphology and pigmentation. In C, a wild-type embryo injected with tfap2a e2i2 MO and tfap2c e3i3 MO, with no visible pigmentation, an absence of yolk extension, a poorly extended tail, and edema ventral to the hindbrain. |
| Genes: | |
|---|---|
| Fish: | |
| Knockdown Reagents: | |
| Anatomical Terms: | |
| Stage: | 1-4 somites |
| Fish: | |
|---|---|
| Knockdown Reagents: | |
| Observed In: | |
| Stage: | Prim-15 |
Reprinted from Developmental Biology, 304(1), Li, W., and Cornell, R.A., Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos, 338-354, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.