- Title
-
DHODH Inhibition Suppresses MYC and Inhibits the Growth of Medulloblastoma in a Novel In Vivo Zebrafish Model
- Authors
- Tsea, I., Olsen, T.K., Polychronopoulos, P.A., Tümmler, C., Sykes, D.B., Baryawno, N., Dyberg, C.
- Source
- Full text @ Cancers
|
|
|
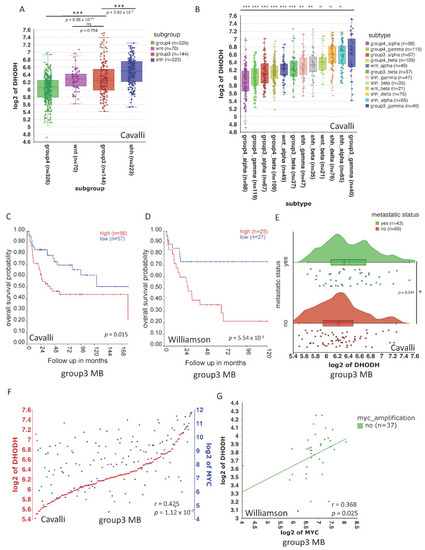
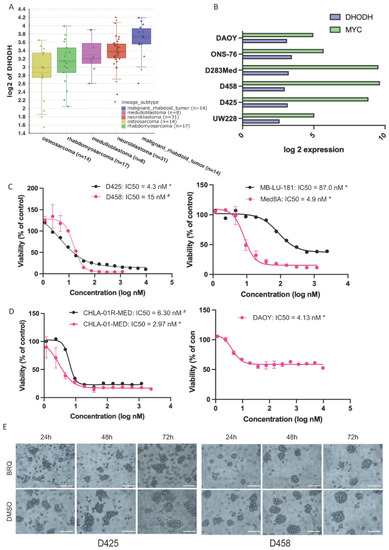
MB cell lines express |
|
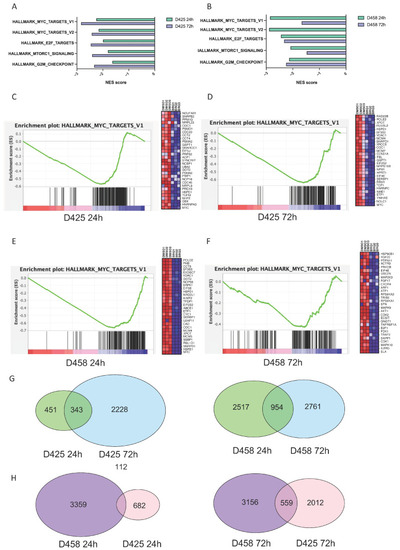
BRQ treatment has a sustained inhibition of MYC target expression. ( |
|
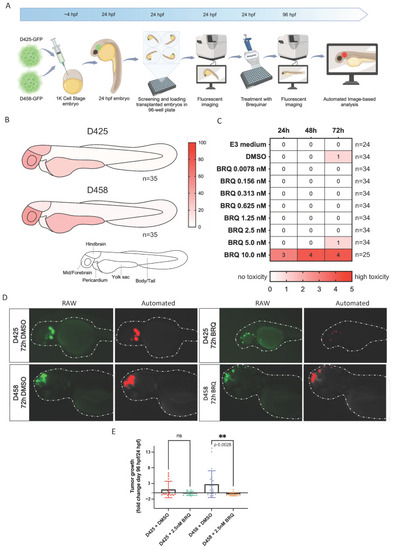
BRQ treatment inhibits tumour growth in a group 3 MB zebrafish xenograft model. ( |