- Title
-
Analysis of a shark reveals ancient, Wnt-dependent, habenular asymmetries in vertebrates
- Authors
- Lanoizelet, M., Michel, L., Lagadec, R., Mayeur, H., Guichard, L., Logeux, V., Séverac, D., Martin, K., Klopp, C., Marcellini, S., Castillo, H., Pollet, N., Candal, E., Debiais-Thibaud, M., Boisvert, C., Billoud, B., Schubert, M., Blader, P., Mazan, S.
- Source
- Full text @ Nat. Commun.
|
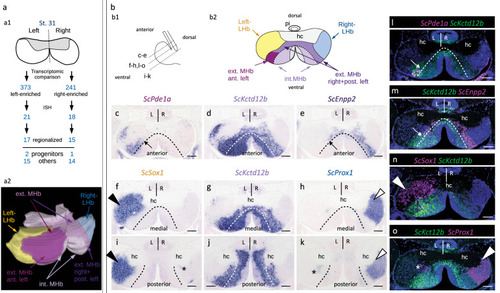
Developing catshark habenulae harbor major asymmetries both in lateral and medial compartments. |
|
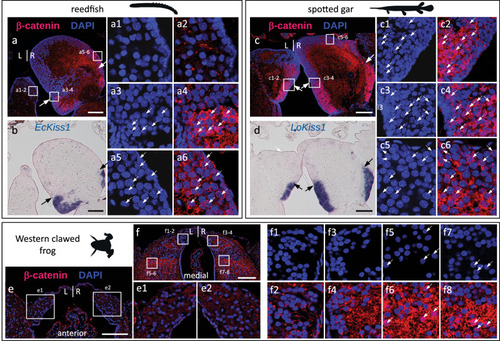
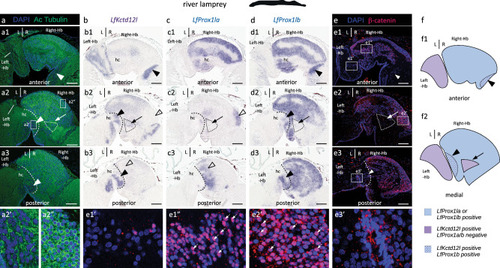
Asymmetries related to those observed in catshark lateral habenulae are present in a lungfish and a polypterid, but undetectable in members of tetrapods and neopterygians. |
|
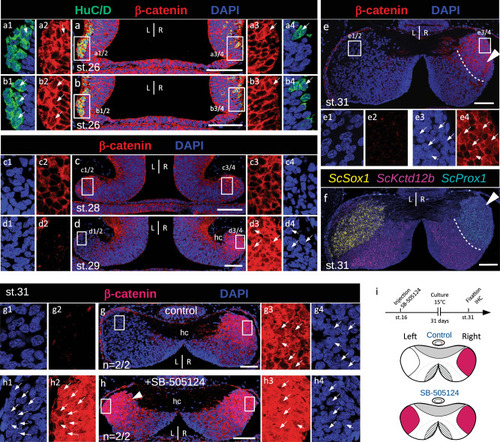
The nuclear β-catenin profile in developing catshark habenulae reveals dynamic, Nodal-dependent asymmetries. |
|
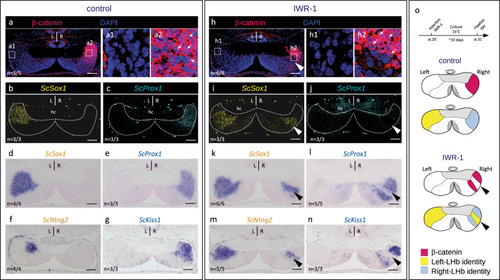
Inhibition of Wnt signaling converts lateral right into lateral left neuronal identities in developing catshark habenulae. |
|
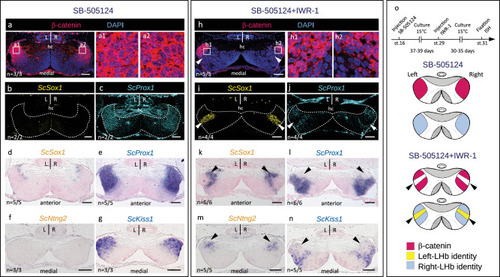
Inhibition of Wnt signaling rescues Left-LHb neuronal identities in the lateral habenulae of SB-505124-treated catshark embryos. |
|
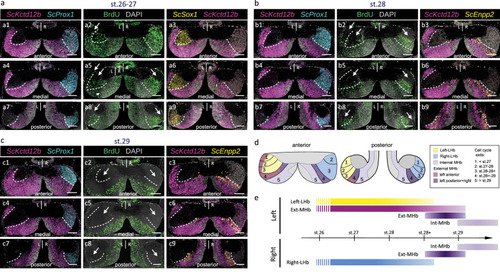
Spatial and temporal regulation of progenitor cell cycle exits in developing catshark habenulae. |
|
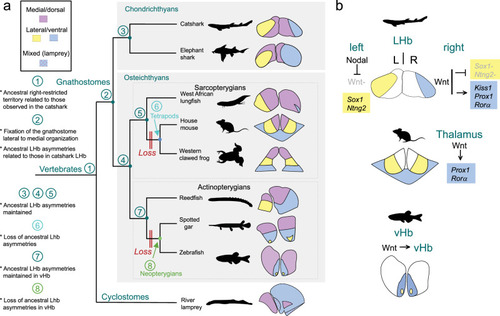
Evolution of nuclear β-catenin asymmetry patterns in the habenulae of jawed vertebrates. |
|
Expressions of |
|
Evolution of habenular asymmetries in jawed vertebrates. |