- Title
-
Integrative mRNA and miRNA Expression Profiles from Developing Zebrafish Head Highlight Brain-Preference Genes and Regulatory Networks
- Authors
- Zhang, S., Yang, J., Xu, J., Li, J., Xu, L., Jin, N., Li, X.
- Source
- Full text @ Mol. Neurobiol.
|
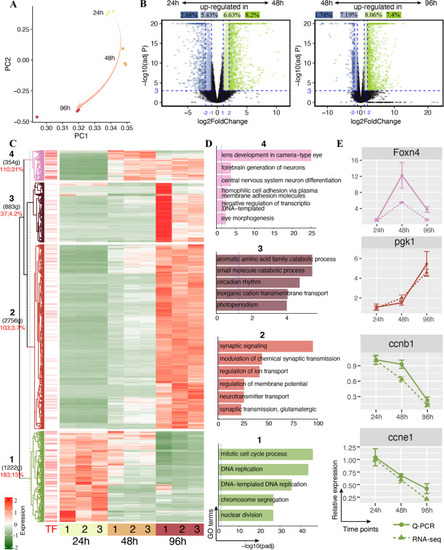
Differentially expressed protein-coding genes (DEGs) in developing zebrafish head transcriptome. |
|
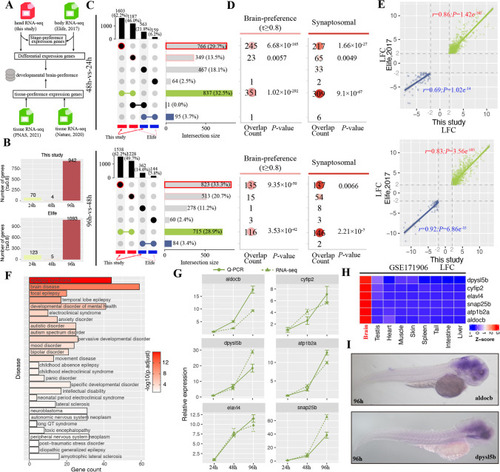
Significant enrichment of brain-preference genes in head transcriptome compared to that in the whole embryo RNA-seq. |
|
|
|
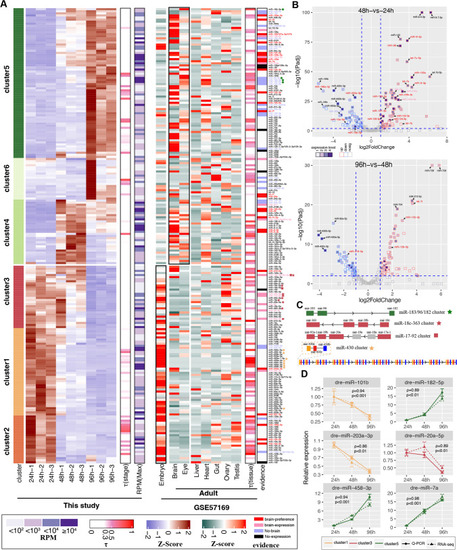
Development-related miRNAs in zebrafish brain. |
|
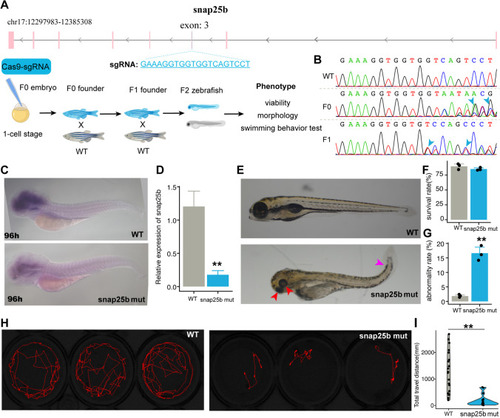
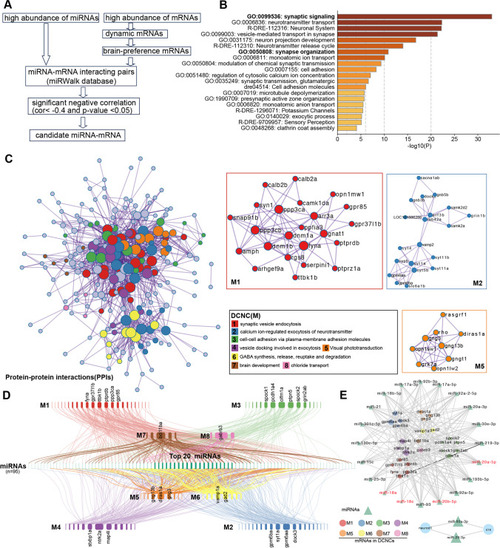
Potential miRNA-mRNA interacted pairs involved in nervous system development. |