- Title
-
Single-cell chromatin profiling reveals genetic programs activating proregenerative states in nonmyocyte cells
- Authors
- Dong, Y., Yang, Y., Wang, H., Feng, D., Nist, E., Yapundich, N., Spurlock, B., Craft, M., Qian, L., Liu, J.
- Source
- Full text @ Sci Adv
|
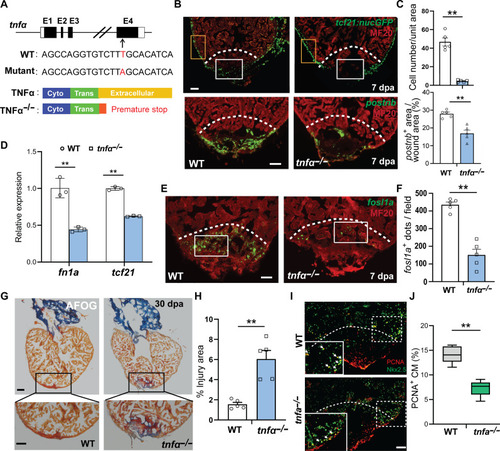
Loss of TNFα function comprises nonCM activation and heart regeneration. ( |
|
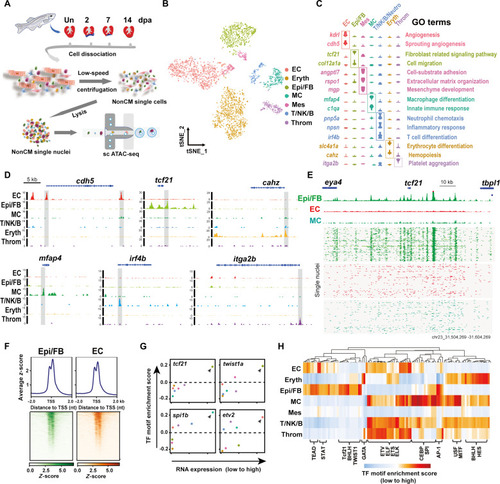
scATAC-seq reveals chromatin accessibility landscape of cardiac nonCMs. ( |
|
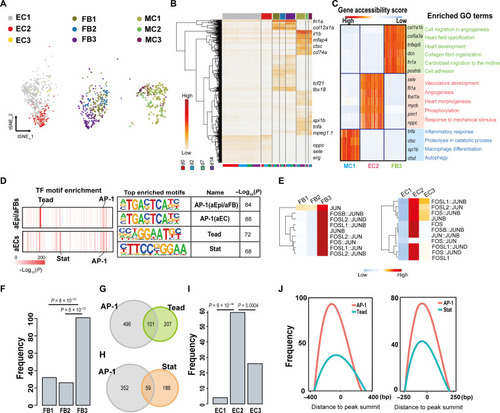
Epi/FBs and ECs show distinct epigenetic features during heart regeneration. ( |
|
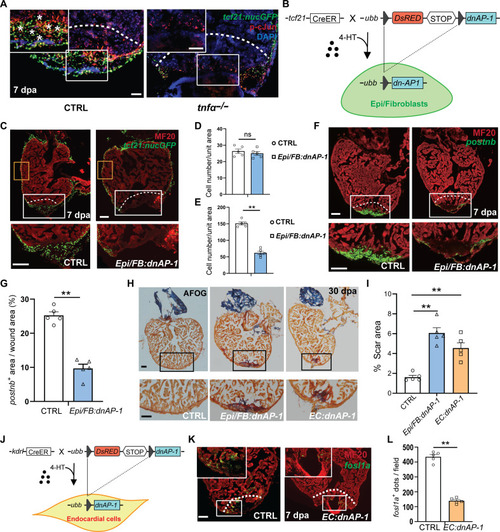
Inhibition of AP-1 activity compromises heart regeneration. ( |
|
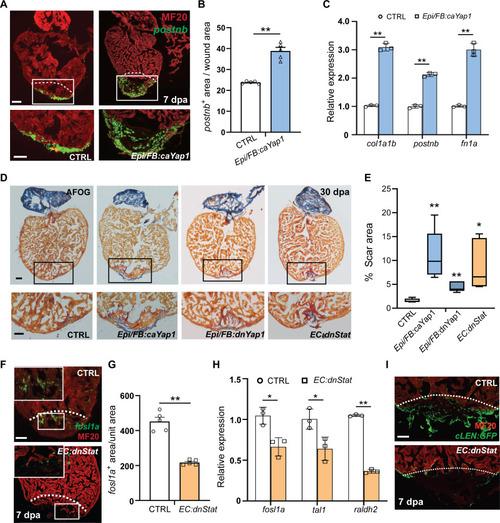
Yap1/Tead activation and inhibition of Stat activity compromises heart regeneration. ( |
|
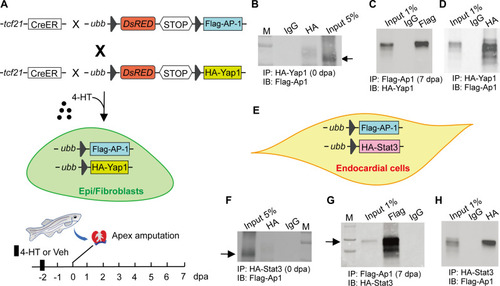
AP-1 interacts with Yap1 and Stat3 in Epi/FBs and ECs, respectively. ( |
|
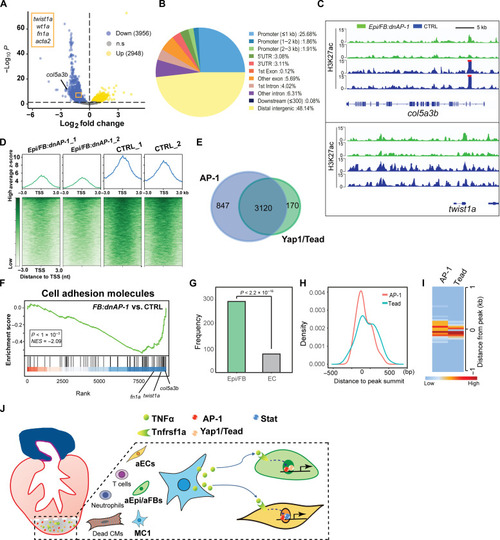
CUT&Tag profiling of H3K27ac reveals nonCM chromatin landscape changes of upon Epi/FB-specific AP-1 inhibition. ( |