Fig. 3.
- ID
- ZDB-FIG-240224-3
- Publication
- Dong et al., 2024 - Single-cell chromatin profiling reveals genetic programs activating proregenerative states in nonmyocyte cells
- Other Figures
- All Figure Page
- Back to All Figure Page
|
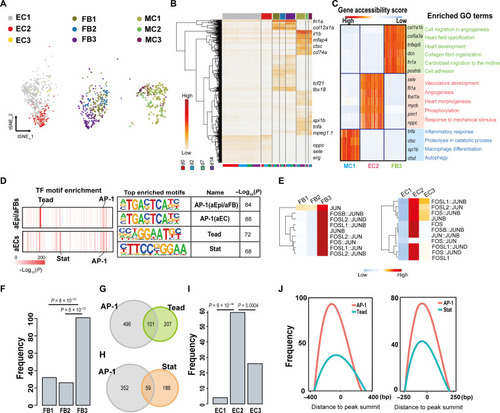
Epi/FBs and ECs show distinct epigenetic features during heart regeneration. ( |