- Title
-
A comparative analysis of gene and protein expression in chronic and acute models of photoreceptor degeneration in adult zebrafish
- Authors
- Kramer, A.C., Carthage, J., Berry, Y., Gurdziel, K., Cook, T.A., Thummel, R.
- Source
- Full text @ Front Cell Dev Biol
|
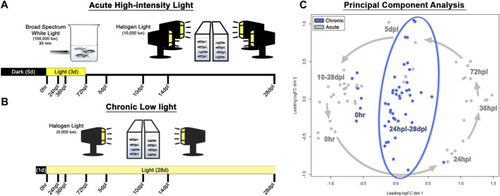
Methods and principal component analysis. |
|
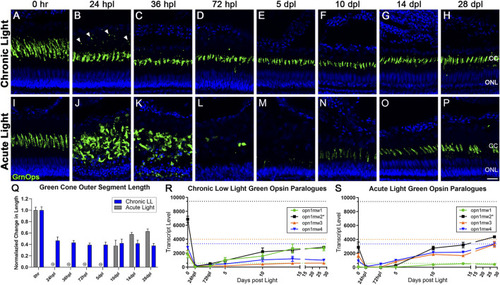
Differential morphology and |
|
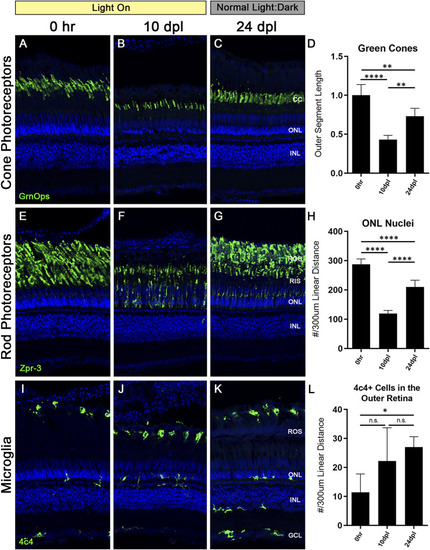
CLL exposure leads to slow rod photoreceptor degeneration, whereas rod photoreceptors are destroyed and replaced in the AL model. Rod PRs were immunolabeled (green) with zpr-3 to mark rod outer segments (ROS) and nuclei were stained blue with TO-PRO-3. |
|
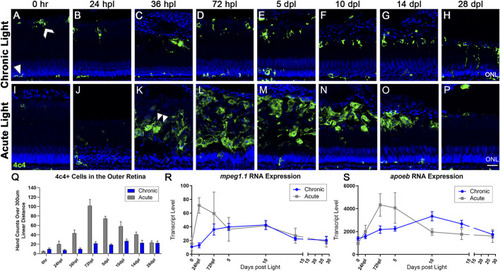
The microglial response to CLL is muted in comparison to AL damage. Anti-4c4 antisera (green) was used to immunolabel microglia and nuclei were stained with TO-PRO-3 (blue). |
|
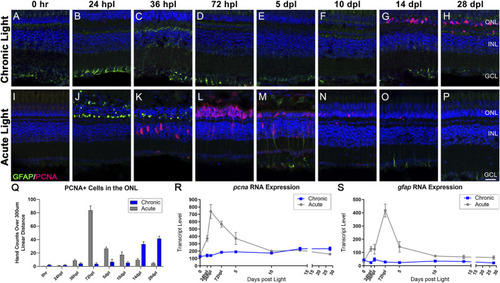
Distinct responses in Müller glia gliosis and proliferation in the CLL and AL models. Müller glia (MG) intermediate filaments were immunolabelled with anti-GFAP antisera (GFAP; green) and cell cycle re-entry was demonstrated with anti-PCNA antisera (PCNA; red). Nuclei were stained with TO-PRO-3 (blue). |
|
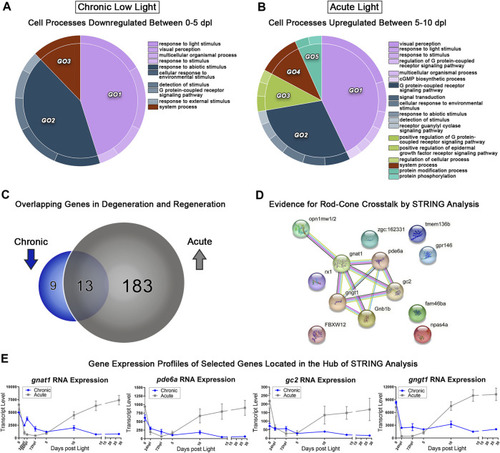
Comparative transcriptomic analysis of photoreceptor degeneration and regeneration. |
|
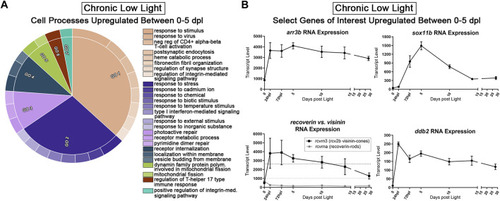
Transcriptomic analysis of early PR degeneration in the CLL Model. |
|
Truncated photoreceptor outer segments recover when zebrafish are transferred back to normal light:dark conditions. Zebrafish were subjected to the CLL lesion protocol for 10 days, then returned to normal light:dark housing conditions and allowed to recover for 14 days. After the 14-day recovery period, retinas were harvested at 24 days post initial light (dpl). |