FIGURE 6
- ID
- ZDB-FIG-231002-407
- Publication
- Kramer et al., 2023 - A comparative analysis of gene and protein expression in chronic and acute models of photoreceptor degeneration in adult zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
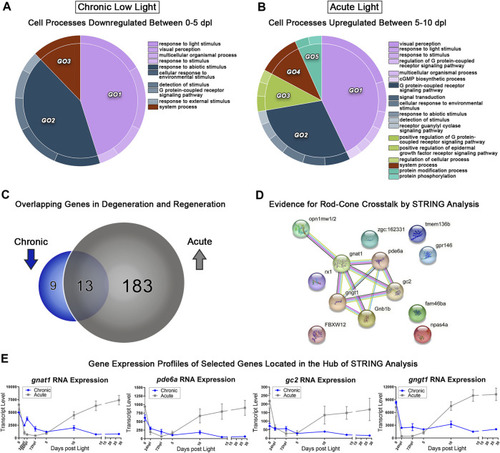
Comparative transcriptomic analysis of photoreceptor degeneration and regeneration. |