- Title
-
Proteomic analysis of zebrafish Folliculogenesis identifies YB-1 (Ybx1/ybx1) as a potential gatekeeping molecule controlling early ovarian Folliculogenesis
- Authors
- Lau, E.S., Zhu, B., Sun, M.A., Ngai, S.M., Ge, W.
- Source
- Full text @ Biol. Reprod.
|
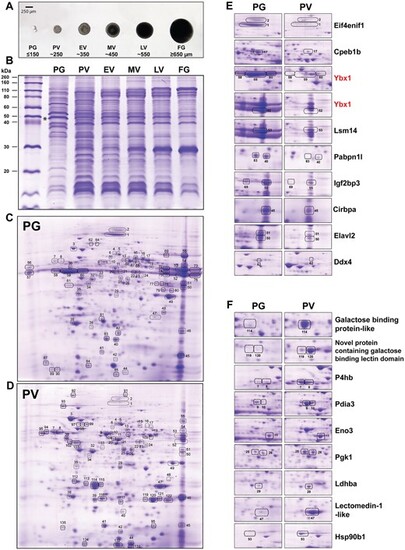
Folliculogenesis in zebrafish and changes in protein profile. (A) Stages of zebrafish follicles. PG (<150 μm); PV (~250 μm); EV (~350 μm); MV (~450 μm); LV (~550 μm); FG (>650 μm). (B) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) protein profiles during folliculogenesis. (C and D) 2D protein profiles of PG and PV stages. Total proteins of PG and PV follicles (80 μg) were used for IEF with IPG strips (7 cm, pH 3–10 linear) respectively. 2D gels were stained with Coomassie blue. The differentially expressed proteins were identified and marked by numbers, and the list of identified proteins is shown in Supplementary Table S2. Some proteins characterized in both PG and PV samples did not exhibit any evident alterations (e.g. #22, 23, 24, 33, 34, and 38) between the two stages, and these proteins can serve as internal controls for loading. (E) Selected proteins down-regulated in PV follicles. (F) Selected proteins up-regulated in PV follicles. Eif4enif1, eukaryotic translation initiation factor 4E nuclear import factor 1 (gi|61,806,649); Cpeb1b, cytoplasmic polyadenylation element-binding protein 1b (gi|82,249,026); Ybx1, Y box-binding protein 1; Lsm14, LSM14 homolog B-like (gi|239,050,742); Pabpn1l, poly(A) binding protein nuclear 1 like (gi|189,521,749); Igf2bp3, insulin-like growth factor 2 mRNA binding protein 3 (gi|18,858,571); Cirbpa, cold inducible RNA binding protein a (gi|62,955,567); Elavl2, ELAV Like RNA Binding Protein 2 (novel protein containing RNA recognition motifs) (gi|94,732,878); Ddx4, DEAD-Box helicase 4 (gi|2,558,535); P4hb, prolyl 4-hydroxylase beta subunit (gi|193,788,703); Pdia3, protein disulfide isomerase A3 (gi|27,881,963); Eno3, enolase 3 (gi|47,551,317); Pgk1, phosphoglycerate kinase 1 (gi|41,388,972); Ldhba, lactate dehydrogenase Ba (gi|28,277,619); Hsp90b1, heat shock protein 90 beta 1 (gi|38,016,165). The two images shown for Ybx1 represent the protein located at the acidic and basic sides of the gel, respectively. |
|
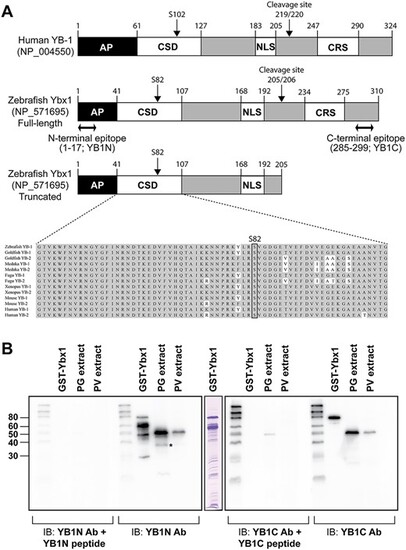
Structure of zebrafish Ybx1 and preparation of anti-Ybx1 antibodies. (A) Domain structure of human YB-1 and zebrafish Ybx1 proteins. AP, AP-rich N-terminal; CSD, cold shock domain; NLS, nuclear localization signal; CRS, cytosol localization signal; YB1N, anti-N-terminal antibody; YB1C, anti-C-terminal antibody. (B) Western blot testing for antibody specificity. YB1N could detect a major band at 48 kDa and a minor band at 36 kDa (asterisk) in PG follicle extract, whereas the YB1C could detect the major band only. |
|
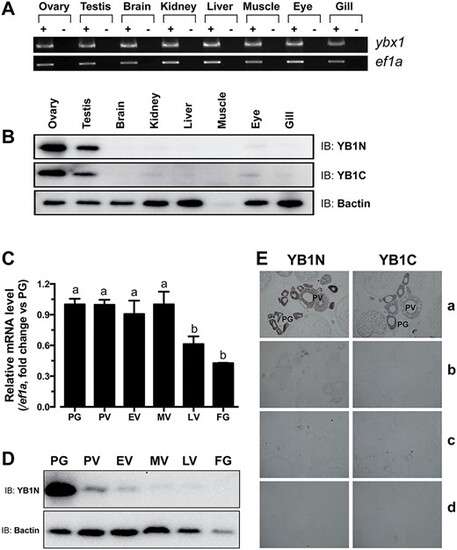
Spatiotemporal expression of Ybx1 mRNA and protein in different tissues and ovarian follicles. (A) Semi-quantitative RT-PCR analysis of ybx1 and ef1a expression in different tissues. +, RT with M-MLV; −, RT without M-MLV. (B) Western blot detection of Ybx1 in different tissues. YB1N, anti-N-terminal antibody; YB1C, anti-C-terminal antibody; Bactin, β-actin. (C) Real-time PCR analysis of ybx1 expression in the follicles of different stages. Housekeeping gene, ef1a, was used as control. Different letters indicate statistical significance (n = 3) (D) Western blot detection of Ybx1 protein in different stages of follicles. (E) Immunohistochemical detection of Ybx1 in follicles. a, with antibodies YB1N or YB1C; b, antibodies neutralized with the antigenic peptides; c, pre-immune sera; d, no antibodies. |
|
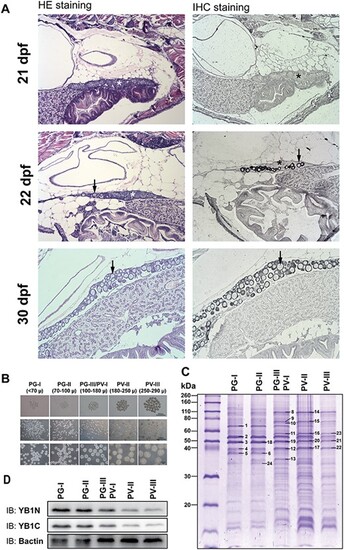
Expression of Ybx1 during ovarian differentiation and follicle activation (PG–PV transition). (A) Immunohistochemical staining for Ybx1 protein during ovarian differentiation. The juvenile fish were sampled at 21, 22, and 30 dpf for histological sectioning and immunohistochemical staining. At 21 dpf, the germ cells or CN oocytes (asterisk) were clustered in cysts without follicle formation. Individual PN follicles (arrow) started to appear at 22 dpf and increased in number at 30 dpf. No signal for Ybx1 could be detected in the cystic CN cells; however, strong signals of Ybx1 appeared in the oocytes once follicles formed during cyst breakdown. (B) Sub-staging of PG and PV follicles. Based on size and morphological features, the PG stage is divided into early PG (PG-I), mid-PG (PG-II), and late PG (PG-III); and PV stage is divided into early PV (PV-I), mid-PV (PV-II), and late PV (PV-III). PG-III and PV-I were difficult to separate and therefore grouped together for analysis. (C) SDS-PAGE analysis for protein profiles during PG–PV transition. The PV-III/PV-I group represented the transitional stage. (D) Western blot for Ybx1 during PG-PV transition. (1) insulin-like growth factor 2 mRNA binding protein 3; (2) Zgc:55673 protein; (3, 18–21) Y-box binding protein 1; (4, 12, 17, 22) actin; (8, 14) heat shock protein 90 kDa beta, member 1; (9, 15) heat shock protein 5; (10) heat shock protein 8; (11, 16, 23) procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide; (13) ribosomal protein lateral stalk subunit P0; (24) glyceraldehyde-3-phosphate dehydrogenase. |
|
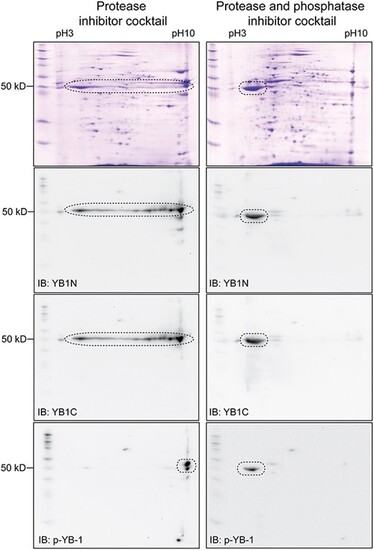
Western blot evidence for multi-site phosphorylation of Ybx1. The protein extract from PG follicles in the presence or absence of phosphatase inhibitors followed by 2D electrophoresis and Western blot analysis with YB1N, YB1C, and anti-p-YB-1. In the absence of phosphatase inhibitor, the signal of Ybx1 protein spanned a wide range of pH from 3 to 10. This pattern disappeared in the presence of phosphatase inhibitors and the signal became concentrated around pH 3. The anti-p-YB-1 detected phosphorylation at S82 site. |
|
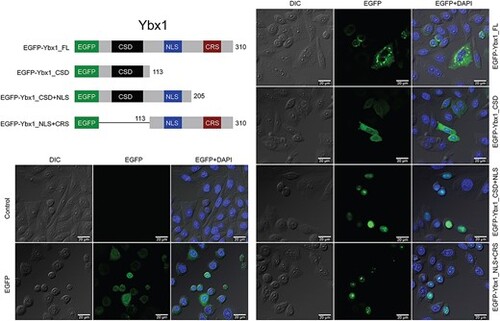
Intracellular localization of EGFP fused with different domains of zebrafish Ybx1. The expression vectors for EGFP-Ybx1 fusion proteins were transfected into the CHO cells followed by confocal microscopic observation. CSD, cold shock domain; NLS, nuclear localization signal; CRS, cytoplasmic retention signal. |
|
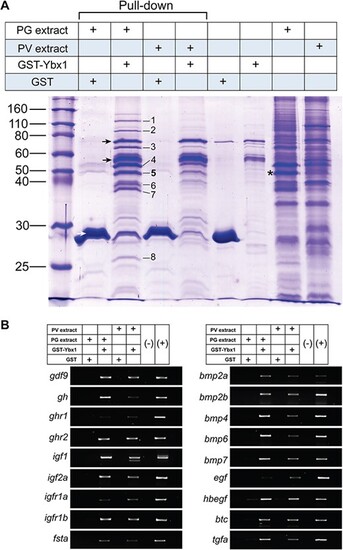
Evidence for Ybx1 binding of mRNAs and identification of its associated proteins in the mRNPs. (A) GST pull-down assay for protein partners of Ybx1 in the mRNPs. The GST-Ybx1 fusion protein was incubated with the extracts from PG or PV follicles followed by pull-down with glutathione Sepharose 4B. The proteins pulled down were subject to SDS-PAGE separation and MS analysis. (1) eukaryotic translation initiation factor 4E nuclear import factor 1 (4E-T) (gi|61,806,649); (2) protein associated with topoisomerase II homolog 2 isoform 1 (gi|125,831,101); (3) insulin-like growth factor 2 mRNA binding protein 3 (gi|18,858,571); (4) ATP-dependent RNA helicase DDX6 (DEAD box protein 6) (gi|68,444,623) and RNA-associated protein 55 (RAP55) (gi|157,423,287); (5) Y box binding protein 1 (YB-1) (gi|29,477,111); (6) novel protein containing RNA recognition motifs (a.k.a. RRM, RBD, or RNP domain) (gi|94,732,878) (7) zygote arrest 1 (gi|56,207,255); (8) polyadenylate-binding protein 2 (PABP2) (gi|189,521,749). Asterisk, Ybx1; black arrow, GST-Ybx1 fusion protein (intact and truncated forms); white arrow, GST. (B) RT-PCR analysis for mRNAs in the mRNPs pulled down with GST-Ybx1 fusion protein. gdf9, growth differentiation factor 9; gh, growth hormone; ghr1 and ghr2, growth hormone receptor 1 and 2; igf1 and igf2a, insulin-like growth factor 1 and 2a; igfr1a and igfr1b, insulin-like growth factor receptor 1a and 1b; fsta, follistatin a; bmp2a, bmp2b, bmp4, bmp6 and bmp7, bone morphogenetic protein 2a, 2b, 4, 6 and 7; egf, epidermal growth factor; hbegf, heparin-binding EGF-like growth factor; btc, betacellulin; tgfa, transforming growth factor α. |
|
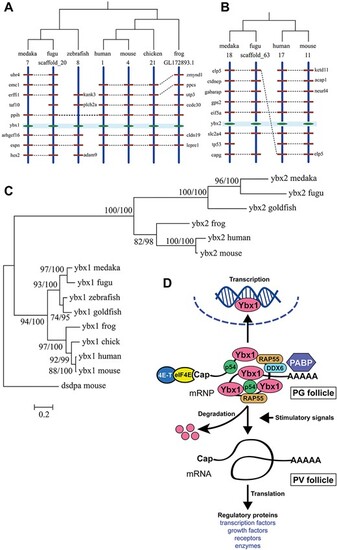
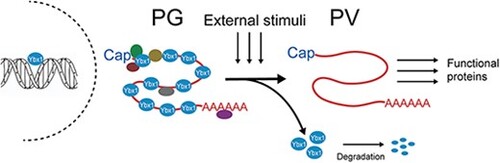
Phylogenetic analysis for YB-1 and YB-2 in zebrafish and the hypothesis for roles of Ybx1 in follicle activation. (A) Chromosome synteny analysis of genes surrounding YB-1 among different organisms. (B) Synteny analysis of genes surrounding YB-2 genes. (C) Topology derived from ML and Bayesian analyses of YB-1 and YB-2 genes (amino acid sequence). The two numbers at a node represents Bootstrap value of ML and Bayesian analysis. The branch lengths are drawn according to ML tree. (D) Hypothetic model for roles of Ybx1 in PG-PV transition. As a key component of mRNPs, Ybx1 binds mRNAs in PG follicles together with other proteins such as PABP, DDX6, RAP55, and p54, stabilizing the messengers while suppressing their translation. At the transition from PG to PV, Ybx1 protein is massively degraded to release the mRNAs from the mRNPs for translation into a variety of regulatory proteins, resulting in follicle activation. |
|
|