- Title
-
Mitochondrial network expansion and dynamic redistribution during islet morphogenesis in zebrafish larvae
- Authors
- Freudenblum, J., Meyer, D., Kimmel, R.A.
- Source
- Full text @ FEBS Lett.
|
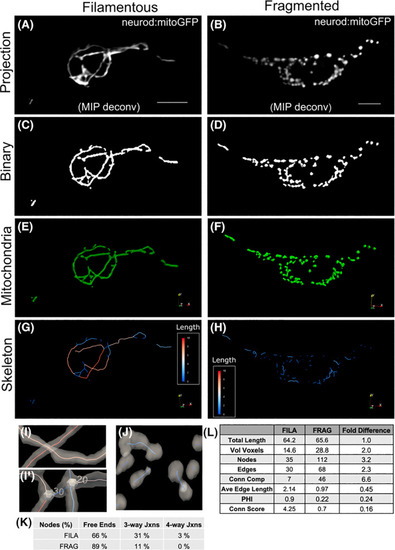
Quantitative analysis of mitoGFP‐labelled mitochondria in secondary islet cells using |
|
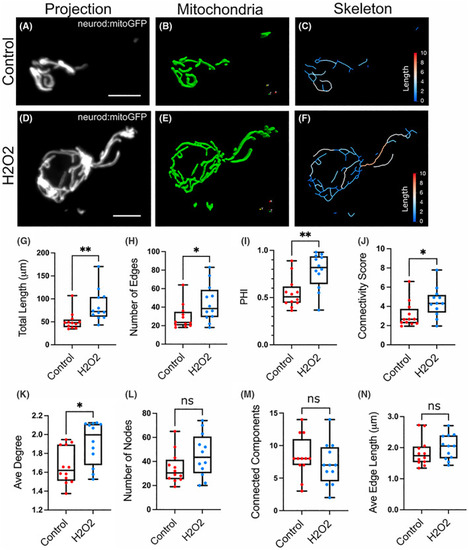
Treatment with H2O2 causes mitochondrial expansion. Representative cells from neuro:mitoGFP larvae at 6 dpf, control (A–C) and H2O2‐treated (D–F). (A, D) Maximum intensity projection of neurod:mitoGFP (grey). Scale bar, 5 μm. Paraview 3D representation of the mitochondrial objects (B, E) and the skeletonised network (C, F) following analysis with |
|
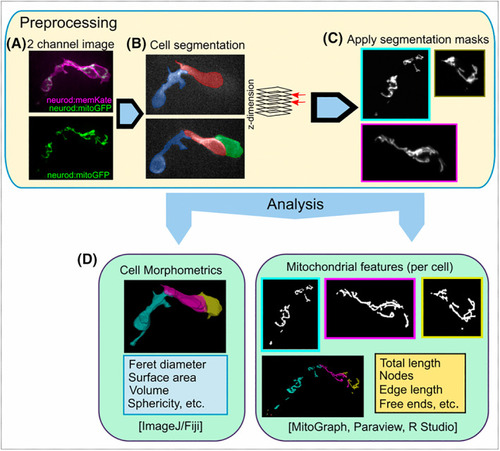
Segmentation of mitochondria in clustering cells. (A) Expression of neurod:memKate in addition to neurod:mitoGFP facilitates segmentation of mitochondria in adjoining cells. Closely opposed mitochondria cannot be assigned to specific cells based on neurod:mitoGFP expression alone (A, bottom). (B) Cell segmentation, shown in two different z‐planes, based on the membrane‐localised memKate signal. (C) By using cell segmentation masks, mitochondria from each cell can be separated. (D) Quantitative analysis of single cell morphology (left) and the corresponding mitochondrial network (right) using open‐source software. |
|
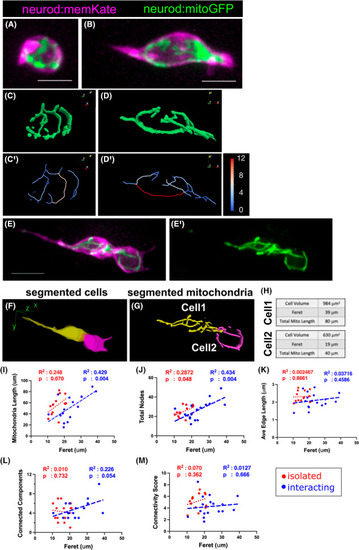
Mitochondrial morphologies of clustering islet cells. (A, B) Maximum intensity projection of image stack of isolated islet cells from neurod:mitoGFP;neurod:memKate transgenics at 7 dpf. Mitochondrial network (C, D) and skeleton (C′, D′) as analysed by |
|
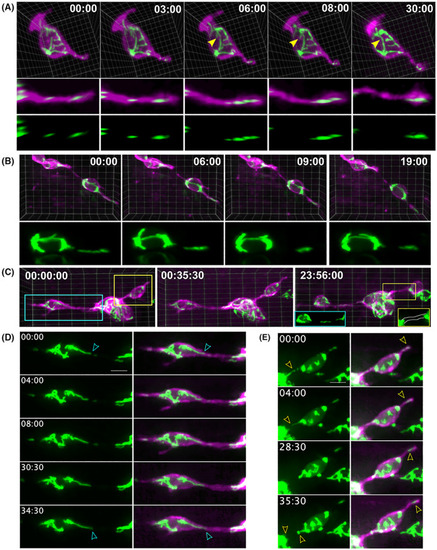
Mitochondrial localisation in relation to cell motility. (A, B) Mitochondria within protrusions move as discrete units (A) or elongated filaments (B). Distally directed mitochondrial displacement over time shown in 3D visualisations of the full cell (A, top), and a rotated view to show the full protrusion length (A, middle), and the mitoGFP signal alone (A, bottom). 3D visualisation of two nearby cells (B) with a filamentous mitochondria shifting distally within a protrusion. (C) 3D view of mitochondria dynamics within clustering cells. When visualised the next day (C, right), the filamentous protrusion has detached from the cell body network (blue inset) and the second cell–cell connection is devoid of mitochondria (yellow inset). (D, E) z‐stack projections highlighting the regions of (A) within the blue box (D) and yellow box (E). (A, B, D, E) Times are as indicated: min : s. (C) Times are as indicated: h : min : s. (D, E) Scale bar, 5 μm. |