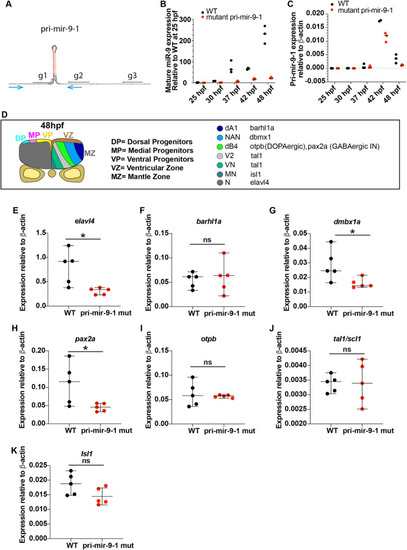
Knocking out the late pri-mir-9-1 preferentially affects neuronal differentiation from medial progenitors. (A) Pri-mir-9-1 hairpin loops with the respective primers used for quantitative PCR annotated as blue arrows (Materials and Methods, mRNA extraction and RT-qPCR; Table S3). Customized guide RNA to delete specifically pri-mir-9-1 are annotated as g1, g2 and g3. Red sequence, miR-9-5′ arm; orange sequence, miR-9-3′ arm; black letters, pre-mir-9; grey sequence, partial sequence of pri-mir-9-1. (B) Taqman RT-qPCR of mature miR-9 from dissected hindbrain at different stages of development, in wild-type conditions (black dots) and deletion of pri-mir-9-1 (red dots), relative to wild-type at 25 hpf. (C) SYBR green RT-qPCR relative quantification of pri-mir-9-1 from dissected hindbrain at different stages of development, in wild-type conditions (black dots) and deletion of pri-mir-9-1 (red dots). Quantification was normalised using β-actin. (D) Schematic of transverse section from zebrafish hindbrain at the level of the otic vesicle at 48 hpf. MZ, mantle zone; VZ, ventricular zone. Within the VZ there are dorsal progenitors (DP), medial progenitors (MP) and ventral progenitors (VP). The schematic shows late neuronal markers expressed in different neuronal cell types in the hindbrain: dA1, dorsal neurons expressing barhl1a; NAN, noradrenergic neurons expressing dbmx1; dB4, GABAergic interneurons expressing pax2a; V2, interneurons expressing tal1; VN, ventral neurons expressing tal1; MN, motor neurons expressing isl1; N, pan neuronal zone expressing elavl4; otpb is localised in the dB4 region but is a marker for dopaminergic neurons. (E-K) SYBR green relative quantification of elavl4 (E), barhl1a (F), dmbx1a (G), pax2a (H), otpb (I), tal1/scl1 (J) and isl1 (K) from dissected hindbrain at 72 hpf, in wild-type conditions (black dots) and deletion of pri-mir-9-1 (red dots). Quantification was normalised using β-actin. Horizontal bars indicate median with 95% confidence intervals. *P<0.05 (Mann-Whitney two-tailed test). (B-C) N=3, each N contain a pool of 10 hindbrain. (E-K) N=5. ns, not significant.
|