- Title
-
A two-step search and run response to gradients shapes leukocyte navigation in vivo
- Authors
- Georgantzoglou, A., Poplimont, H., Walker, H.A., Lämmermann, T., Sarris, M.
- Source
- Full text @ J. Cell Biol.
|
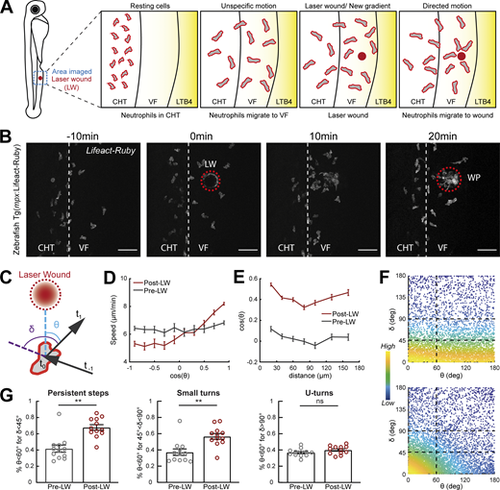
Gradients in vivo cause a directional bias on speed and persistence and small reorientations towards the source. Analysis of neutrophil migration responses to gradients in zebrafish. (A) Scheme showing two-photon LW chemotaxis assay in zebrafish. Resting neutrophils at the CHT are stimulated to exit the VF after LTB4 addition. Laser wounding in the VF at 30 min after LTB4 addition redirects neutrophil motion. (B) Time-lapse sequence of two-photon confocal image projections, showing fluorescent neutrophils in 3 dpf Tg(mpx:Lifeact-Ruby) transgenic larva, migrating pre- and post-LW (red dashed line). LW occurs at 0 min. Scale bar = 50 μm. (C) Scheme for angles δ and θ. δ is the angle between the speed vector of three successive timepoints (t0–t−1, t1–t0). θ is the angle between the speed vector (t1–t0) and the vector connecting the neutrophil geometrical center to the closest point of the LW perimeter. (D) Cell speed in relation to the cosine of θ for pre- and post-LW. n = 343–1,283 cell steps per bin pre-wound, n = 236–3,168 cell steps per bin post-wound. (E) Cell cosine of θ in relation to the distance from the closest point of the WP. n = 517–863 cell steps per bin for pre-wound, n = 444–1,272 cell steps per bin for post-wound. (F) Scatter plot of cell δ in relation to θ, color-coded for the density of points, for pre- (top) and post-LW (bottom). Black dashed lines indicate the groups of data points taken for analysis for G. Data shown from 12 larvae. (G) Percentage of cell persistent steps (δ < 45°), small turns (45° < δ < 90°) and U- turns (δ > 90°), respectively, with θ < 60°. Wilcoxon matched-pairs signed rank test, **, P = 0.0024 (persistent steps), P = 0.0068 (small turns). (D, E, and G) Data from 12 larvae in each case. |
|
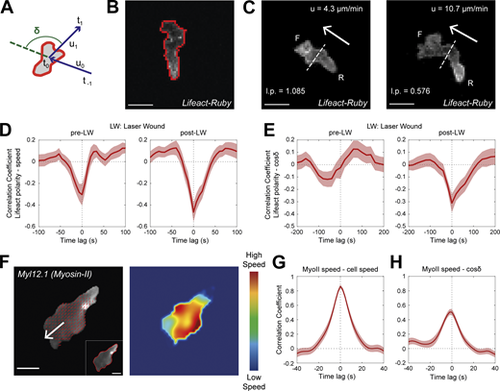
Rear Lifeact distribution reports phases of fast motion and fast actin flows. (A) Scheme of a neutrophil, indicating the definition of angle δ, between the velocity vectors (u) between three successive timepoints (t−1, t0, t1). (B) Outline (red line) of an example of segmented neutrophil within a Tg(mpx:Lifeact-Ruby) transgenic larva. Scale bar = 10 μm. (C) Examples of neutrophil Lifeact distribution with an indication of cell speed (u) and Lifeact polarity (l.p.) at the same timepoint, for laser wounding. Arrows indicate the direction of motion. Dashed lines indicate the automated separation of the front (F) and rear (R) part of cell. Scale bar = 10 μm. (D and E) Temporal cross-correlation between Lifeact polarity and speed (D) or between Lifeact polarity and cosine of δ (E), pre- (left) and post-LW (right). Average from n = 21 migrating cells, from 9 larvae. Mean and SEM are shown. (F) Left: Representative neutrophil within a Tg(actb1:myl12.1-eGFP) transgenic larva migrating towards a mechanical wound; white arrow indicates the vector of speed; red arrows indicate the velocity vector fields of Myosin-II retrograde flow; inset (bottom right) indicates the segmented outline of the neutrophil. Scale bar = 5 μm. Right: Heatmap of the speed of the Myosin-II retrograde flow. Color bar indicates low- and high-speed values. (G) Temporal cross-correlation between Myosin-II retrograde flow speed and neutrophil speed. (H) Temporal cross-correlation between Myosin-II retrograde flow speed and neutrophil cosine of δ. (G and H) n = 25 cells, from 6 larvae. Mean and SEM are shown. |
|
A two-stage response to newly encountered gradients in vivo. (A) Time-lapse sequence of two-photon confocal image projections showing a neutrophil (white) in a Tg(mpx:Lifeact-Ruby) zebrafish larva, migrating pre- and post-LW. Cell movement started at 1.5-min after laser wounding for this example cell. White arrows indicate the speed vector. Red arrows indicate the side of cell with higher Lifeact abundance. Scale bar = 10 μm. (B and C) Neutrophil Lifeact polarity, speed, cosine of θ, and cosine of δ, in relation to time, respectively. Time sequence was synchronized based on time of the LW (B) or the time that each individual neutrophil began to move post-LW (C). (B)n = 21 cells, from 9 larvae. (C)n = 18 cells, from 9 larvae. (D) Neutrophil track straightness for motion completed within 3 (top) or 6 (bottom) min before and after beginning of neutrophil movement (BM). n = 18 cells, from 9 larvae. Mean and SEM are shown. Wilcoxon matched-pairs signed rank test, **, P = 0.0040 (3 min), P = 0.0019 (6 min). |
|
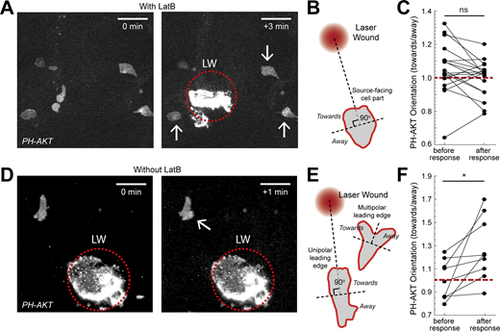
Actin dynamics are required for gradient detection. (A and D) Neutrophils in a Tg(mpx:PHAKT-EGFP) before and after their response to laser wounding (LW; red dotted circle), in the presence (A) or absence (D) of LatB. Arrow points to cells showing local increases in PIP3 intensity. Scale bar = 25 μm. (B) Scheme that shows segmentation of immobilized neutrophils in two parts, facing the source or the opposite direction. (C) Average neutrophil PIP3 orientation (towards/away) in LatB-treated neutrophils, before and after response to gradient. n = 18 cells from 6 larvae. Two-tailed paired t test. (E) Scheme that shows the segmentation of neutrophils, with either unipolar or multipolar leading edge into two parts, one facing the source or the opposite direction. (F) Average neutrophil PIP3 orientation before and after response to gradient (towards/away). n = 9 cells from 7 larvae. Two-tailed paired t test, *, P = 0.0176. (C and F) Red dashed line denotes orientation equal to 1. |
|
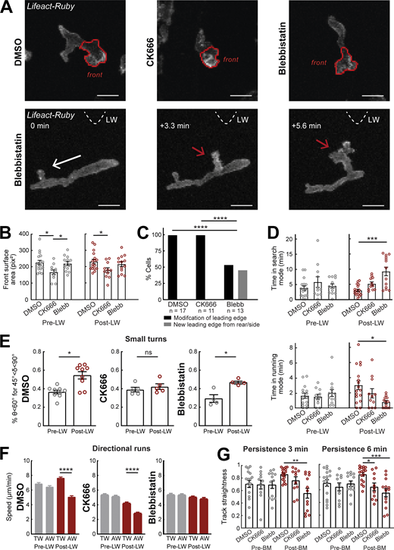
Differential contributions of Arp2/3 and Myosin-II in gradient responses. (A) Top row: Two-photon time-lapse image projections showing neutrophils in Tg(mpx:Lifeact-Ruby) zebrafish larvae treated with DMSO, CK666, and Blebbistatin, where the cell front has been segmented automatically (red lines). Bottom row: Example neutrophil treated with Blebbistatin, with new leading edge forming at the cell lateral/rear towards the LW (red arrow). White arrow indicates the cell’s direction of migration. Triangle indicates the direction of LW (not in the field of view). Scale bar = 10 μm. (B) Average surface area of the cell front part with DMSO, CK666, and Blebbistatin treatments. One-way ANOVA with Dunnett’s multiple comparisons test, * P = 0.0138 (DMSO-CK666 pre-LW), P = 0.0437 (CK666-Blebbistatin pre-LW), P = 0.0379 (DMSO-CK666 post-LW). (C) Percentage of cells that generate a new leading edge from the rear or the side of their body, for the treatments of DMSO, CK666, and Blebbistatin, respectively. Chi-square test, ****, P < 0.0001. (D) Average time that cells spend in search (front Lifeact enrichment) and running (rear Lifeact enrichment) phase, for the treatments of DMSO, CK666 and Blebbistatin, respectively, for pre- (left) and post-LW (right). Kruskal-Wallis test with Dunn’s multiple comparison test, ***, P = 0.0005 for time in search mode, *, P = 0.0124 for time in running mode. (E) Percentage of cells with persistent steps (θ < 60° for δ < 45°). n = 9 larvae for DMSO, n = 5 larvae for CK666, and n = 4 larvae for Blebbistatin. Wilcoxon matched-pairs signed rank test, *, P = 0.0195 for DMSO, P = 0.0216 for Blebbistatin. (F) Mean instantaneous speed for movement towards (TW) or away (AW) from the wound, for pre- and post-LW. n = 1,334–2,292 cell steps per condition for DMSO (9 larvae), n = 1,056–1,616 cell steps per condition for CK666 (5 larvae), and n = 818–1,826 cell steps per condition for Blebbistatin (4 larvae). Kruskal-Wallis test with Dunn’s multiple comparison test, ****, P < 0.0001 for both DMSO and CK666. (G) Persistence of cells for 3 min (left) and 6 min (right) after neutrophil movement for DMSO, CK666, and Blebbistatin. One-way ANOVA with Dunnett’s multiple comparisons test, **, P = 0.0004 (3 min, DMSO-Blebbistatin), *, P = 0.0154 (6 min, DMSO-CK666), ***, P = 0.0002 (6 min, DMSO-Blebbistatin). (B–D and G) Data from n = 17 cells from 8 larvae for DMSO, n = 11 cells from 5 larvae for CK666, and n = 13 cells from 4 larvae for Blebbistatin. Mean and SEM are shown. |
|
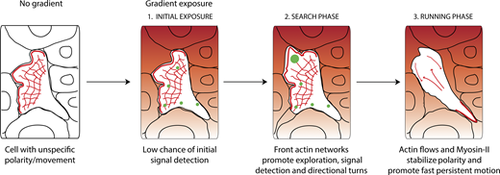
Neutrophils navigate interstitial gradients in vivo through a search and run strategy. Model for neutrophil gradient sensing in vivo. Before encountering the gradient, neutrophils have a pre-established, unspecific polarity, and movement. Upon gradient exposure, neutrophils have a low chance of resolving gradient direction without active actin dynamics, therefore a search phase is required. During this phase, front actin network expansion via Arp2/3 promotes exploration and gradient sampling (illustrated by green spots), culminating in small turns towards the gradient source. Subsequently, cells switch to a running phase, whereby fast actin flows and Myosin-II contractility enforce fast and persistent motion towards the gradient. |