- Title
-
Functional Differentiation of BMP7 Genes in Zebrafish: bmp7a for Dorsal-Ventral Pattern and bmp7b for Melanin Synthesis and Eye Development
- Authors
- Dong, X.R., Wan, S.M., Zhou, J.J., Nie, C.H., Chen, Y.L., Diao, J.H., Gao, Z.X.
- Source
- Full text @ Front Cell Dev Biol
|
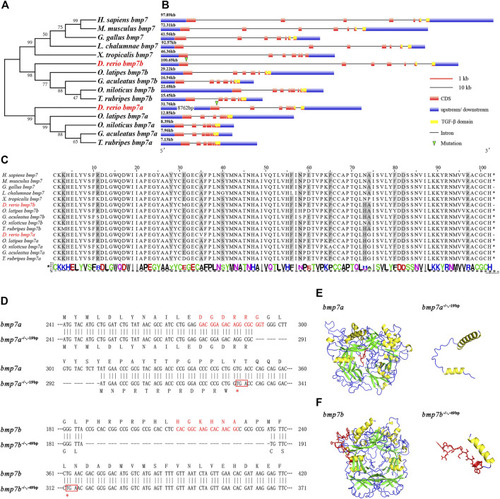
Phylogenetic analysis, conservative sequence, and protein structure prediction of BMP7 genes. |
|
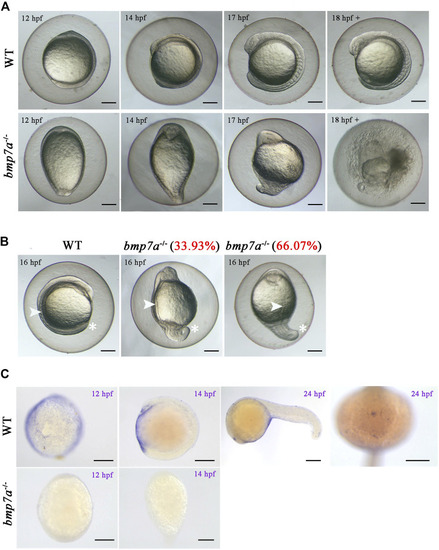
Observation of the homozygous lethal phenotype and gene expression in EXPRESSION / LABELING:
PHENOTYPE:
|
|
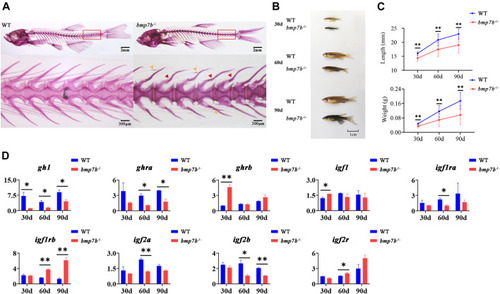
The bone and growth phenotype, and gene expression differences between the wild-type and PHENOTYPE:
|
|
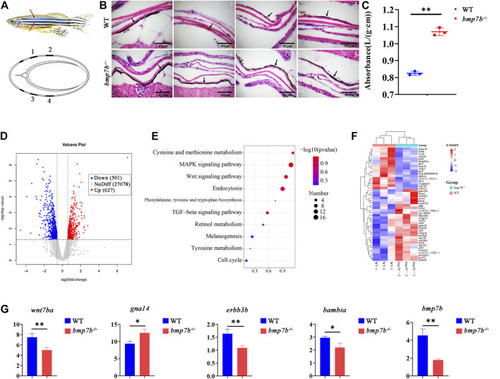
The skin structure, melanin content and gene expression differences between the wild-type and EXPRESSION / LABELING:
PHENOTYPE:
|
|
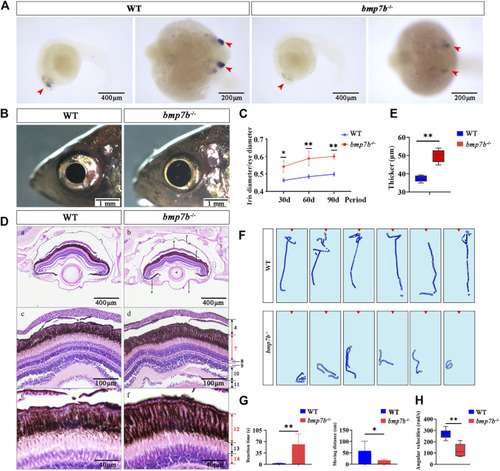
The eye structure, feeding behavior and gene expression differences between the wild-type and |
|
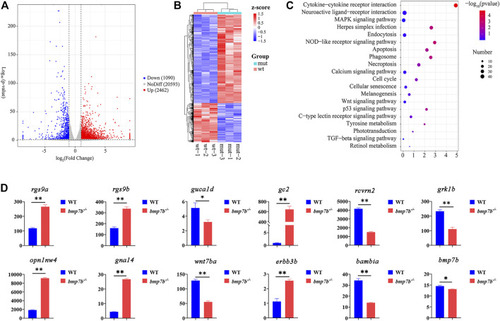
The transcriptome and gene qRT-PCR analysis of eye tissues between the wild-type and EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|