- Title
-
Identification of an evolutionarily conserved domain in Neurod1 favouring enteroendocrine versus goblet cell fate
- Authors
- Reuter, A.S., Stern, D., Bernard, A., Goossens, C., Lavergne, A., Flasse, L., Von Berg, V., Manfroid, I., Peers, B., Voz, M.L.
- Source
- Full text @ PLoS Genet.
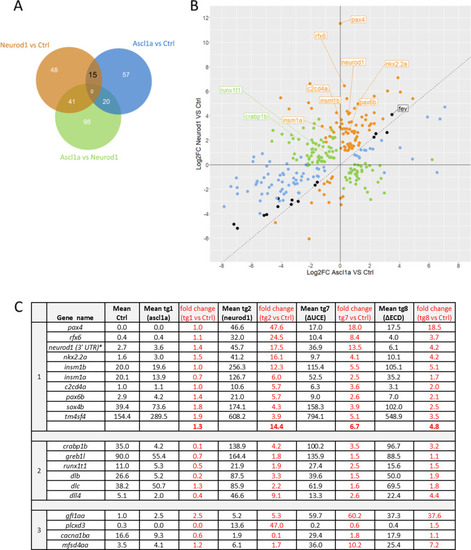
|
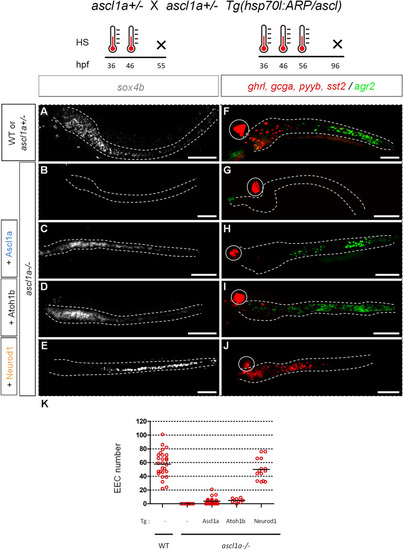
Upper part: scheme of the crossings and timing of the heat shocks (HS). Lower part: |
|
|
|
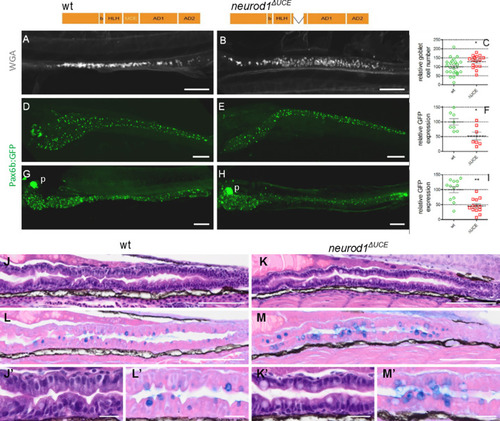
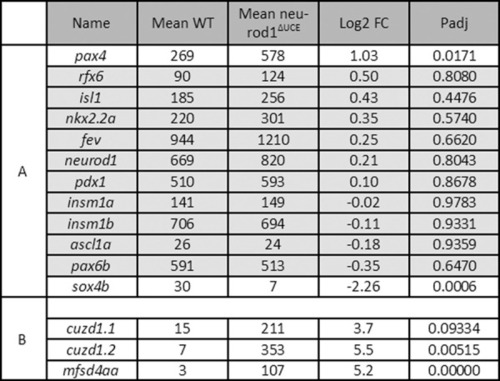
PHENOTYPE:
|
|
|
|
|
|
|
|
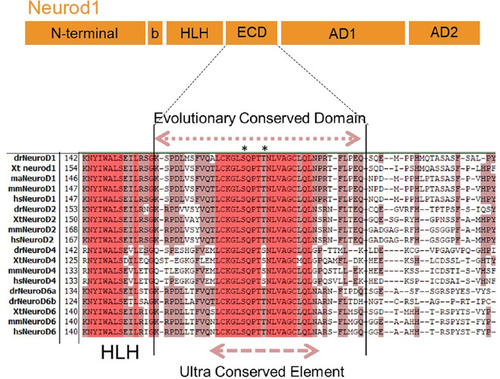
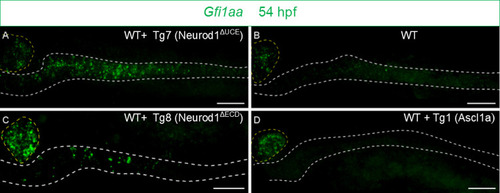
FISH performed with the |
|
|
|
|
|
|
|
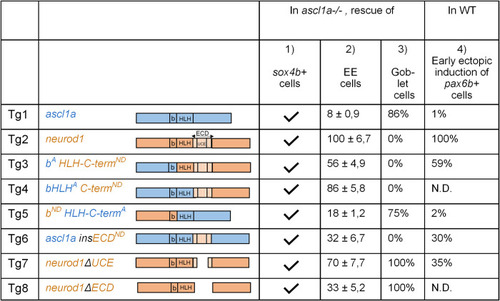
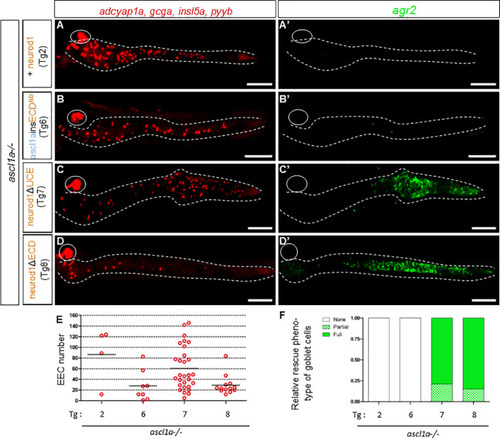
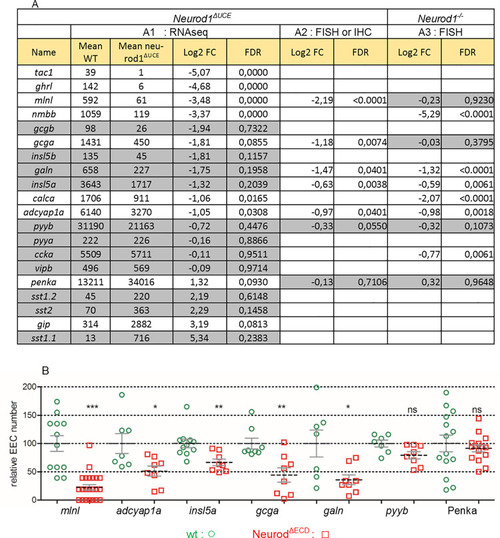
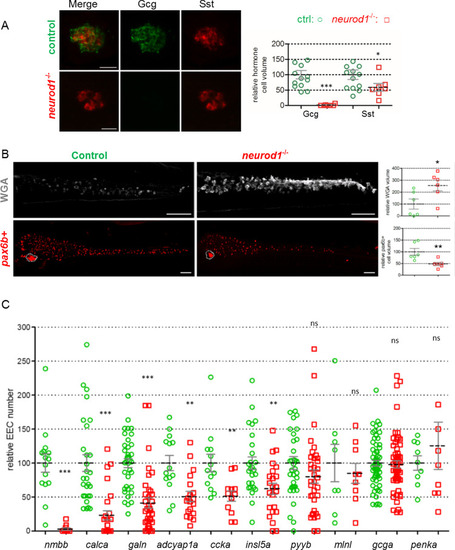
PHENOTYPE:
|