- Title
-
Bi-allelic variants in IPO8 cause a connective tissue disorder associated with cardiovascular defects, skeletal abnormalities, and immune dysregulation
- Authors
- Ziegler, A., Duclaux-Loras, R., Revenu, C., Charbit-Henrion, F., Begue, B., Duroure, K., Grimaud, L., Guihot, A.L., Desquiret-Dumas, V., Zarhrate, M., Cagnard, N., Mas, E., Breton, A., Edouard, T., Billon, C., Frank, M., Colin, E., Lenaers, G., Henrion, D., Lyonnet, S., Faivre, L., Alembik, Y., Philippe, A., Moulin, B., Reinstein, E., Tzur, S., Attali, R., McGillivray, G., White, S.M., Gallacher, L., Kutsche, K., Schneeberger, P., Girisha, K.M., Nayak, S.S., Pais, L., Maroofian, R., Rad, A., Vona, B., Karimiani, E.G., Lekszas, C., Haaf, T., Martin, L., Ruemmele, F., Bonneau, D., Cerf-Bensussan, N., Del Bene, F., Parlato, M.
- Source
- Full text @ Am. J. Hum. Genet.
|
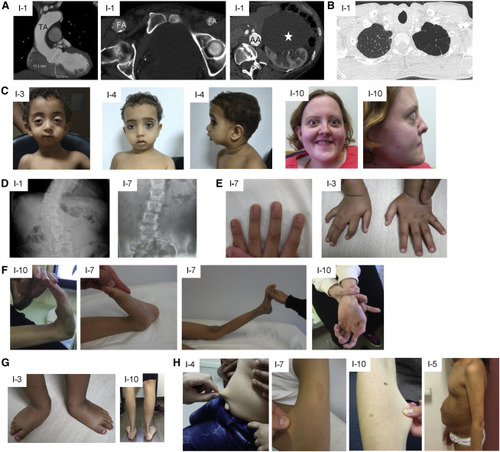
Clinical features of affected individuals (A) CT scan showing dilated aortic root and thoracic aorta (TA), calcified asymmetric large femoral arteries (FA), calcified and dilated abdominal aorta (AA), and large renal cyst (white star) in individual 1. (B) CT scan showing emphysema of the apex of the left lung in individual 1. (C) Proptosis, micrognathia, and hypertelorism in individuals 3, 4, and 10. (D) X-ray showing scoliosis in individuals 1 and 7. (E) Arachnodactyly in individuals 3 and 7. (F) Hyperlaxity of small and large joints in individuals 10 and 7. (G) Pes planus and talipes equinovarus in individuals 10 and 3, respectively. (H) Skin hyperextensibility in individuals 4, 7, and 10 and umbilical hernia in individual 5. |
|
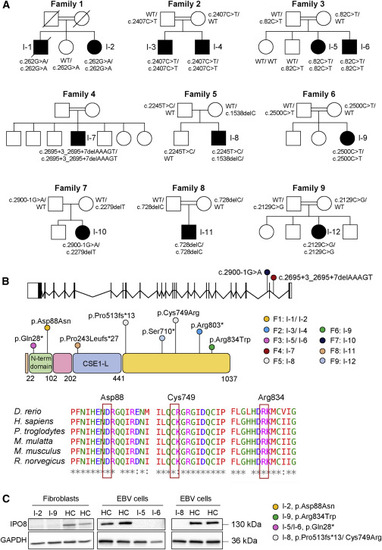
Molecular features of identified variants (A) Pedigrees of families 1–9. (B) Schematic representation of IPO8 gene (25 exons, 24 introns) and importin 8 protein (1,037 amino acids), which features the importin N-terminal domain and the CSE1-like domain, containing tandem HEAT repeats and a RanGTP-binding motif. IPO8 variants identified in this study as well as their location are shown. Protein sequence alignments of IPO8 orthologs are provided for each affected residue. (C) Importin 8 expression in fibroblasts or EBV cells derived from individuals 2, 5, 6, 8, and 9. GAPDH served as loading control. |
|
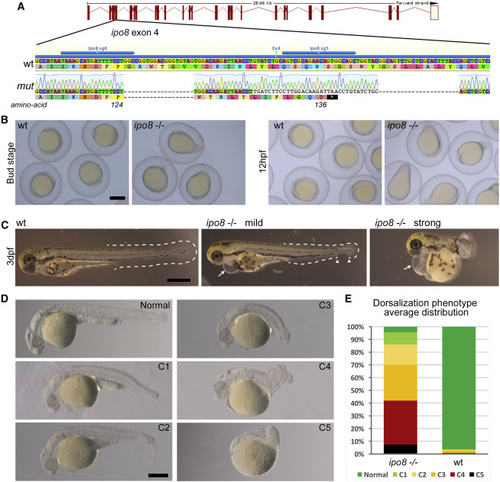
Zebrafish ipo8 mutants show a range of dorsalization phenotypes (A) Schematics of CRISPR/Cas9-mediated gene disruption at the ipo8 genomic locus. The sgRNAs (sg5 and sg6, blue arrows) targeted exon 4. Compared to the wild-type (WT) sequence, the mutated allele from the founder fish (mut) displayed two deletions of 18 and 22 bases and an insertion of 34 bp generating a frameshift from amino acid 124 and a premature STOP codon after 136 amino acids. (B) Bright field pictures of bud (10 hpf) and early somite stages (12 hpf) embryos showing the elongating shape of the ipo8−/− embryos compared to WT controls. Scale bar represents 500 μm. (C) Bright field pictures of 3 dpf larvae presenting the variable penetrance of the ipo8−/− phenotype (mild and strong) compared to WT controls. The tail fin is circled with dashed lines when visible. Gaps are highlighted by arrowheads. Arrows point at heart edema. Scale bar represents 200 μm. (D) Nomarski pictures depicting the different classes of the dorsalization phenotype at 24 hpf as described in Mullins et al.26 (from severe C5 to mild C1) in the ipo8−/− mutants compared to the normal phenotype of a WT embryo. Scale bar represents 200 μm. (E) Quantification of the distribution of the dorsalization classes in ipo8−/− mutants (5 clutches, n = 903) compared to WT clutches (4 WT clutches, n = 503). Average values in ipo8−/− mutants were 7.3% C5, 34.6% C4, 28.0% C3, 16.0% C2, 9.6% C1, and 4.5% normal, and average values in WT controls were 0% C5, 0.8% C4, 1.6% C3, 1.1% C2, 0% C1, and 96.5% normal. PHENOTYPE:
|
|
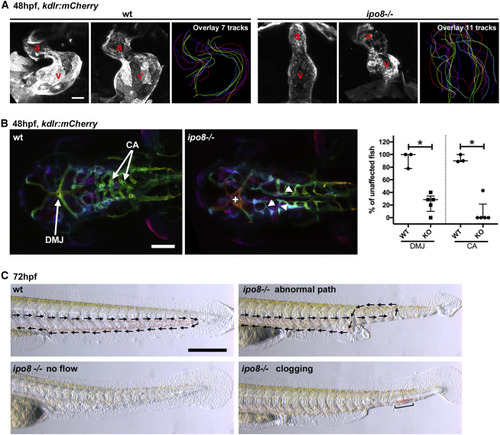
Ipo8 deficiency causes cardiovascular defects in zebrafish (A) Defects in heart chamber formation exemplified by two maximal projections of confocal Z stacks of the hearts of WT and ipo8−/− 48 hpf embryos carrying the kdrl:mCherry transgene to label endothelial cells. For each line, the third panel shows the overlays of the outlines of the 7 (WT) and 11 (ipo8−/−) analyzed hearts. a, atrium; v, ventricle. Scale bar represents 30 μm. (B) Maximal projections of depth color-coded confocal Z stacks of the head vessels of WT and ipo8−/− embryos at 48 hpf, and quantifications of the percentage of fish showing dorsal midline junction (DMJ) defects and central arteries (CA) differentiation defects. Twenty-three WT and 30 ipo8−/− embryos from 3 and 5 clutches, respectively, were analyzed. Median and IQR are shown. p values were calculated by Mann Whitney test (∗p < 0.05). +, abnormal DMJ; arrowheads, abnormal CA. Scale bar represents 100 μm. (C) Maximal projections of time-lapses of Nomarsky imaging of the tails of 3 dpf larvae highlighting absence of blood circulation (no flow) or defects in blood vessel patterning (abnormal path and clogging) in ipo8−/− mutants. Arrows indicate flow direction. Scale bar represents 200 μm. |
|
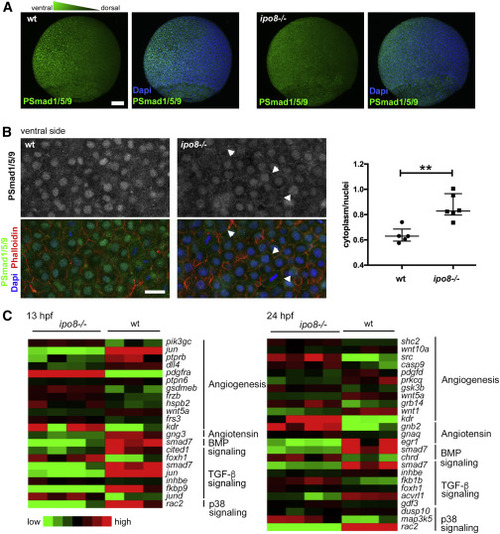
pSMAD nuclear translocation defects in ipo8−/− embryos highlight impaired TGF-β/BMP signaling (A) Maximal projections of confocal Z stacks of WT and ipo8−/− embryos labeled for pSmad1/5/9 (green) and DAPI (blue) showing well-defined ventral to dorsal gradient of pSmad5 in WT gastrulating zebrafish embryos (left panel) but not in the ipo8−/− embryos (right panel). Scale bar represents 100 μm. (B) Confocal images of the ventral sides of WT and ipo8−/− embryos labeled for pSmad1/5/9 (green), phalloidin (red), and DAPI (blue). Arrowheads highlight the membrane localization of the pSmad staining in the ipo8−/− mutants. Scale bar represents 20 μm. Dot plots representative of three experiments show quantifications of the ratio of pSmad1/5/9 cytoplasmic signal over nuclear signal; 5 WT and 6 ipo8−/− embryos were analyzed. Median and IQR are shown. p values were calculated by Mann Whitney test (∗∗p < 0.01). (C) Heatmaps of differentially expressed genes between WT and ipo8−/− embryos at 13 and 24 hpf. Three to four biological replicates are shown per group. The top enriched gene ontology term for biological process is highlighted for each cluster. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

Unillustrated author statements PHENOTYPE:
|