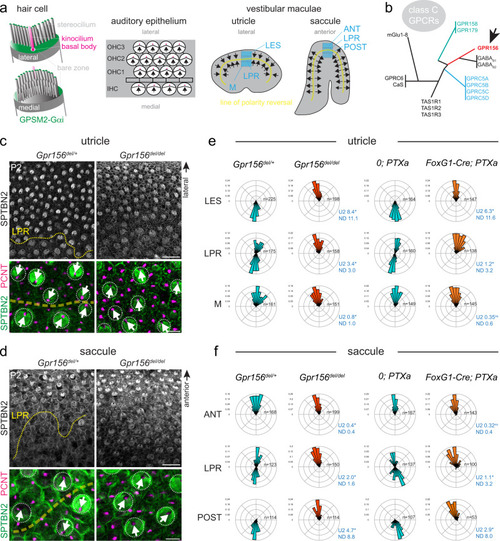
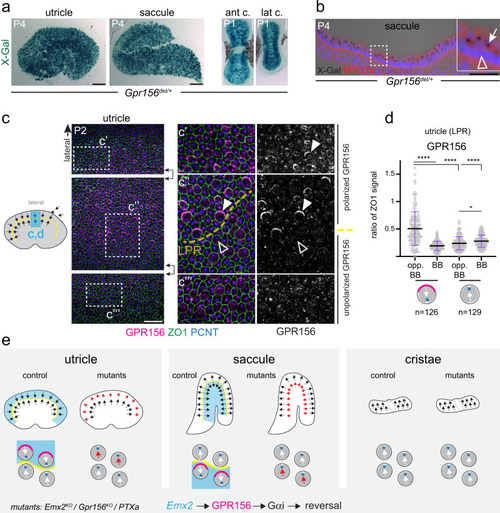
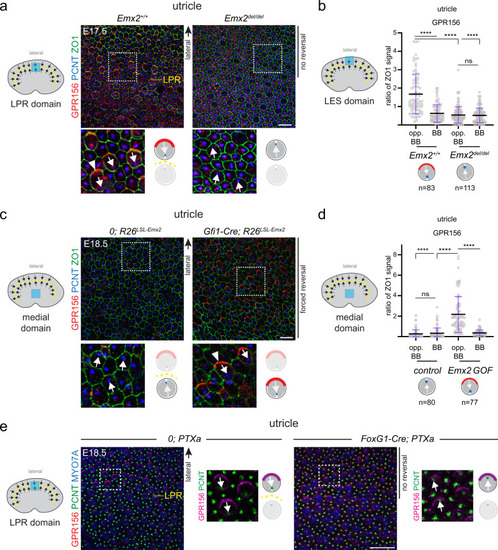
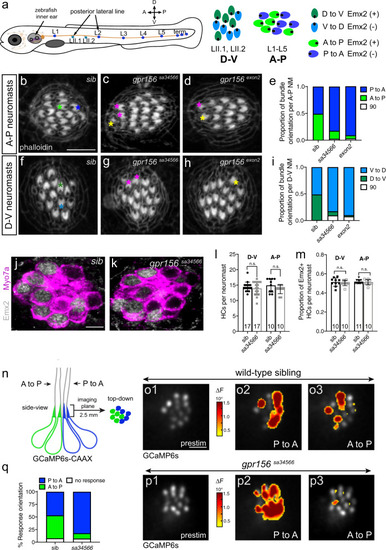
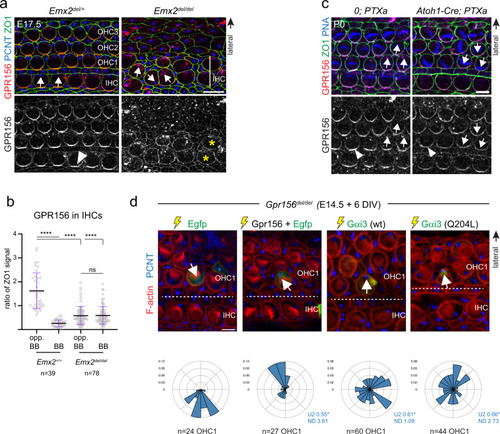
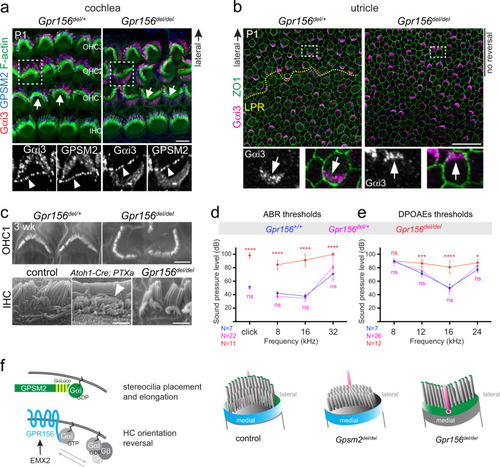
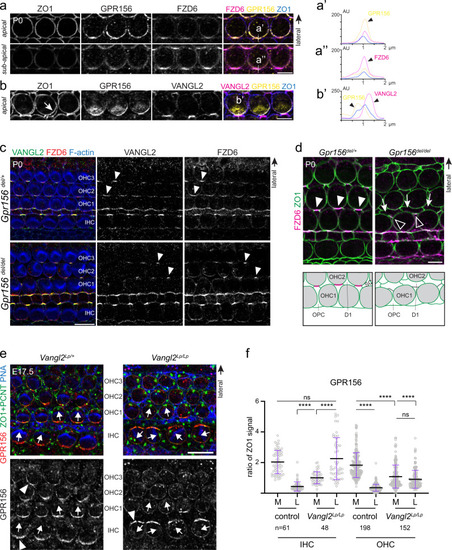
- Title
-
EMX2-GPR156-Gαi reverses hair cell orientation in mechanosensory epithelia
- Authors
- Kindt, K.S., Akturk, A., Jarysta, A., Day, M., Beirl, A., Flonard, M., Tarchini, B.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
|
|
|
|
|