- Title
-
Systematic identification of A-to-I RNA editing in zebrafish development and adult organs
- Authors
- Buchumenski, I., Holler, K., Appelbaum, L., Eisenberg, E., Junker, J.P., Levanon, E.Y.
- Source
- Full text @ Nucleic Acids Res.
|
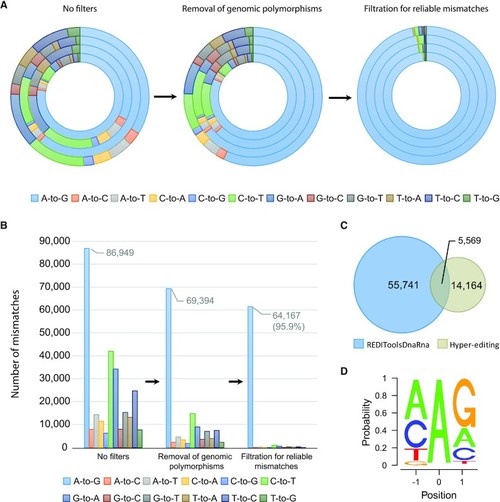
Schematic representation of the process for identifying RNA editing sites in zebrafish. ( |
|
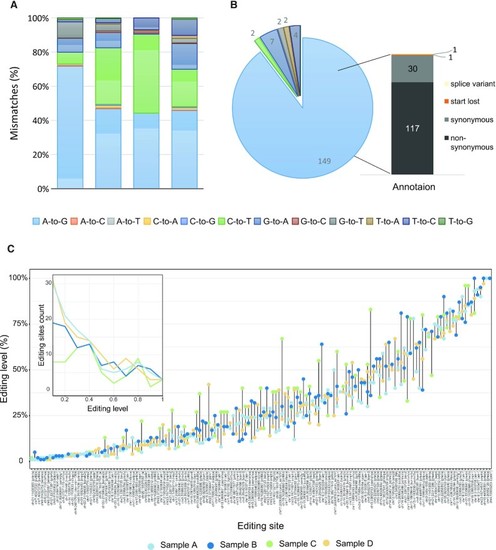
A-to-I editing sites within coding sequences. ( |
|
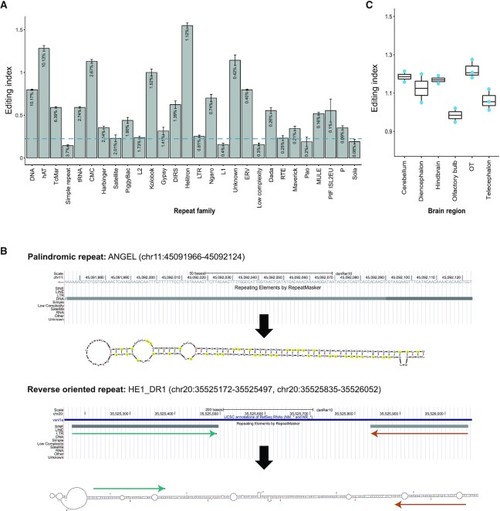
Editing in zebrafish repeats. ( |
|
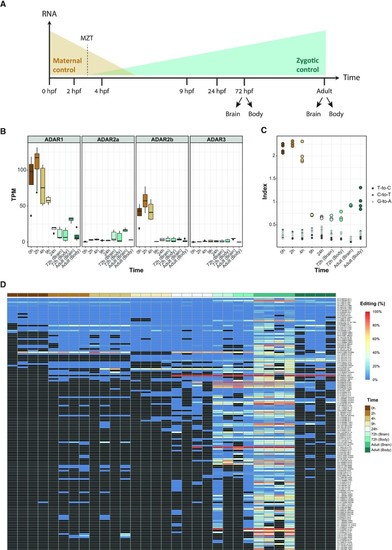
RNA editing during zebrafish development. ( |
|
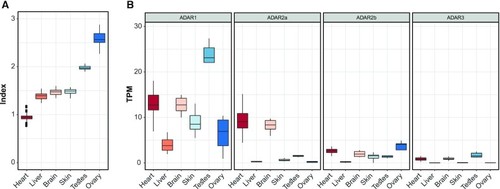
RNA editing across zebrafish tissues. ( |