- Title
-
Loss of splicing factor IK impairs normal skeletal muscle development
- Authors
- In Ka, H., Seo, H., Choi, Y., Kim, J., Cho, M., Choi, S.Y., Park, S., Han, S., An, J., Chung, H.S., Yang, Y., Kim, M.J.
- Source
- Full text @ BMC Biol.
|
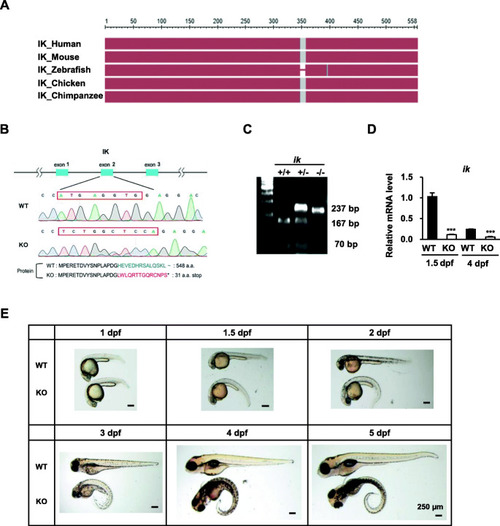
CRISPR/Cas9-mediated |
|
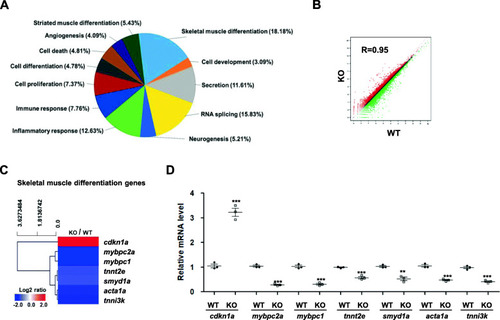
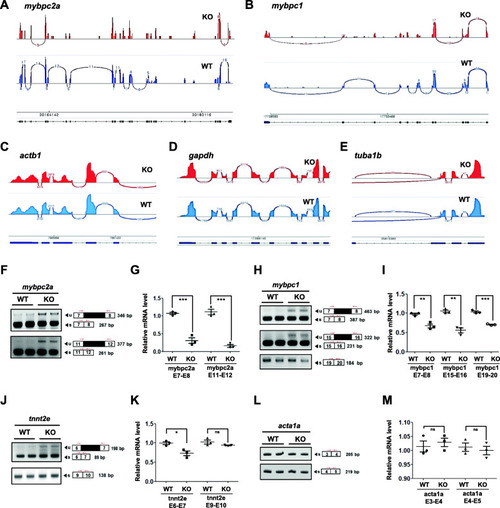
RNA-seq analysis of EXPRESSION / LABELING:
PHENOTYPE:
|
|
PHENOTYPE:
|
|
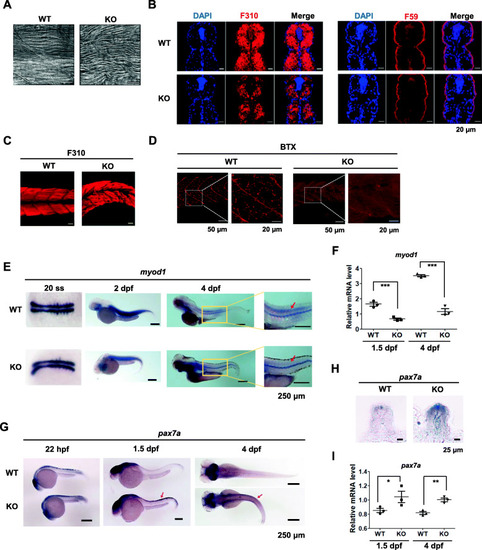
Fast-twitch muscle fibers are impaired in EXPRESSION / LABELING:
PHENOTYPE:
|
|
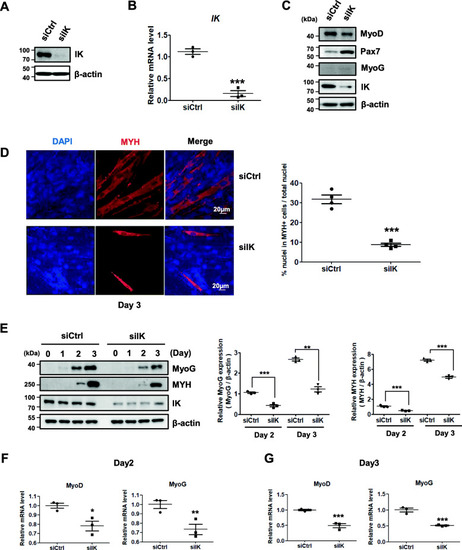
IK-depleted myoblasts have a reduced ability to form normal myotubes. |
|
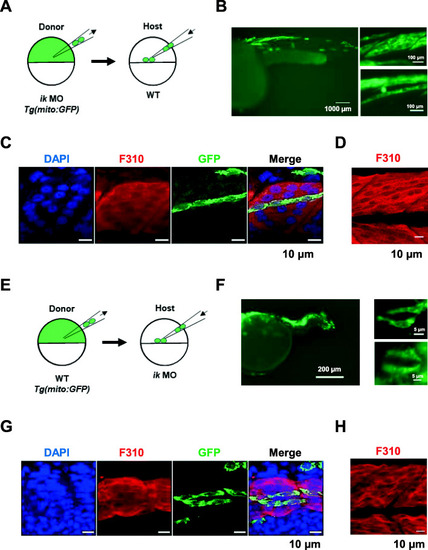
IK functions in a non-cell-autonomous manner in muscle precursors in zebrafish. |
|
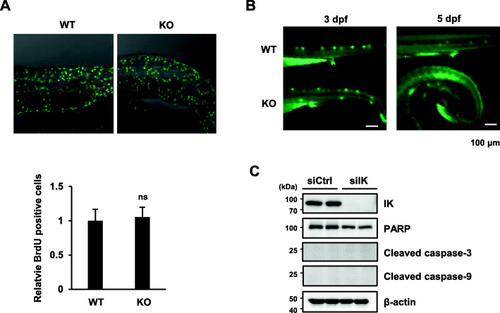
Myoblast proliferation and apoptosis was not affected in |