- Title
-
Impact of low-frequency coding variants on human facial shape
- Authors
- Liu, D., Alhazmi, N., Matthews, H., Lee, M.K., Li, J., Hecht, J.T., Wehby, G.L., Moreno, L.M., Heike, C.L., Roosenboom, J., Feingold, E., Marazita, M.L., Claes, P., Liao, E.C., Weinberg, S.M., Shaffer, J.R.
- Source
- Full text @ Sci. Rep.
|
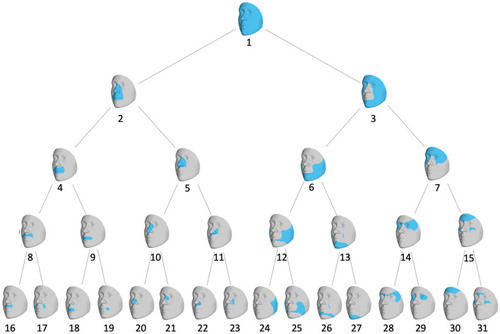
Hierarchical clustering of facial shape. Global-to-local facial segmentation obtained using hierarchical spectral clustering. Segments are colored in blue. The highest-level segment representing the full face was split into two sub-segments, and this bifurcation process was repeated until a five-level hierarchy comprising 31 segments was formed. |
|
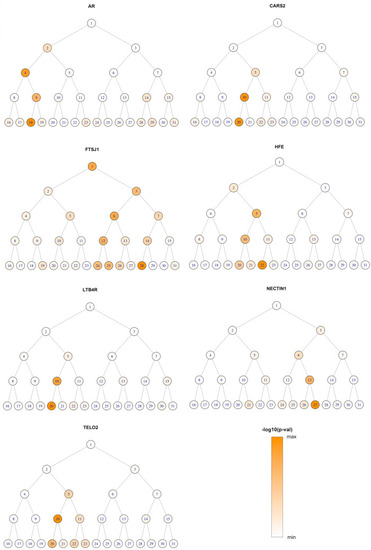
Composite Manhattan plot showing results across 31 facial modules. Manhattan plot showing the position of genes on the x axis and MultiSKAT p-values on the y axis. A total of 31 points are plotted for each gene, representing their p-values in each of the 31 modules. The red horizontal line indicates the significance threshold (3.3 × 10–7). The associated facial modules and the corresponding p-value for each gene that surpassed the threshold (marked as green dots) are shown above the Manhattan plot. The numbers to the bottom left of the facial images indicate the module identifiers in Fig. |
|
Module-wide association results for significant genes. For each gene, the –log10 p |
|
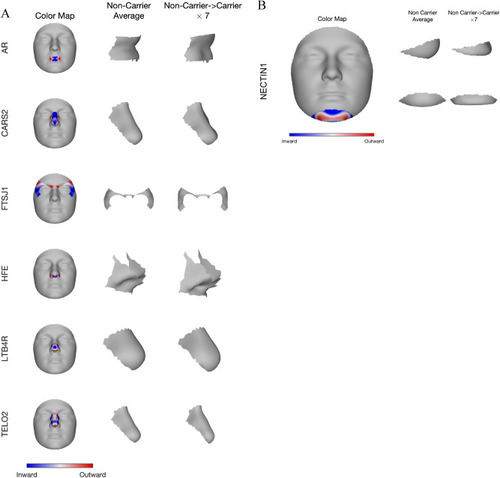
Phenotypic effect of the seven identified genes in their top associated module. Blue and red indicate a local shape depression and protrusion, respectively, due to carrying the low-frequency variants in the gene. ( |
|
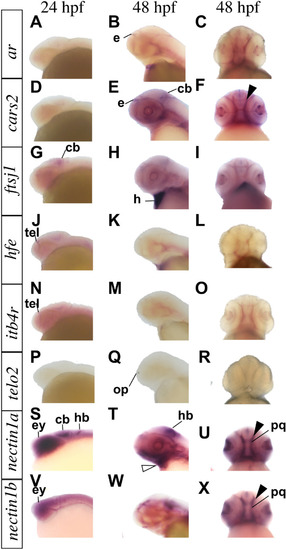
Whole-mount RNA in situ hybridization demonstrating genes expression in zebrafish. Genes expression pattern in lateral and ventral views at the indicated embryonic stages as hours per fertilization (hpf). |
|
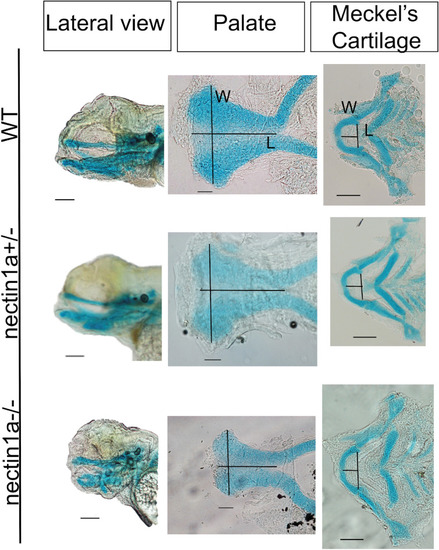
Alcian blue images for PHENOTYPE:
|