- Title
-
Cohesin Components Stag1 and Stag2 Differentially Influence Haematopoietic Mesoderm Development in Zebrafish Embryos
- Authors
- Ketharnathan, S., Labudina, A., Horsfield, J.A.
- Source
- Full text @ Front Cell Dev Biol
|
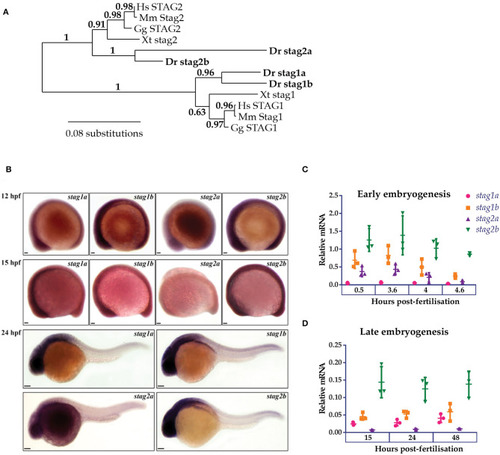
Phylogenetic analysis and embryonic expression of Stag paralogues. |
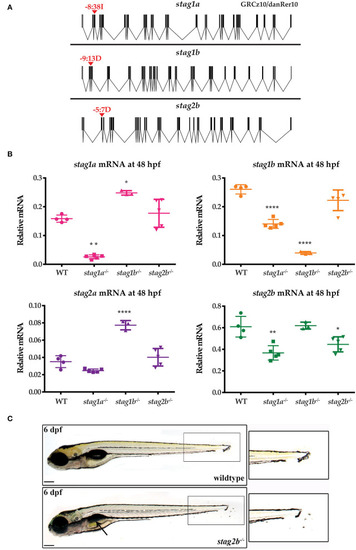
|
Generation of zebrafish |
|
|
|
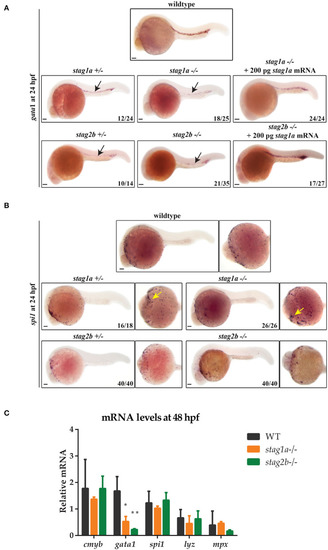
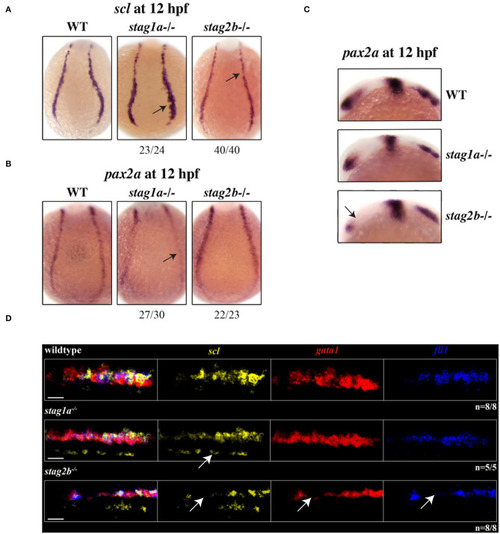
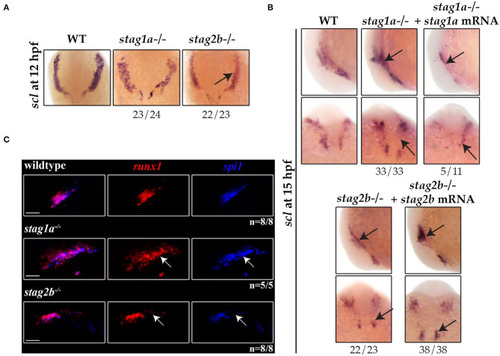
EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
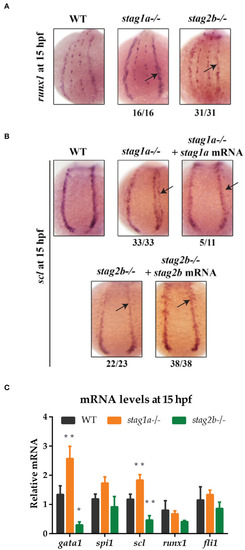
Hypothetical model explaining the effects of Stag1a and Stag2b loss on primitive erythropoiesis. In |

Unillustrated author statements PHENOTYPE:
|