- Title
-
In vivo High-Content Screening in Zebrafish for Developmental Nephrotoxicity of Approved Drugs
- Authors
- Westhoff, J.H., Steenbergen, P.J., Thomas, L.S.V., Heigwer, J., Bruckner, T., Cooper, L., Tönshoff, B., Hoffmann, G.F., Gehrig, J.
- Source
- Full text @ Front Cell Dev Biol
|
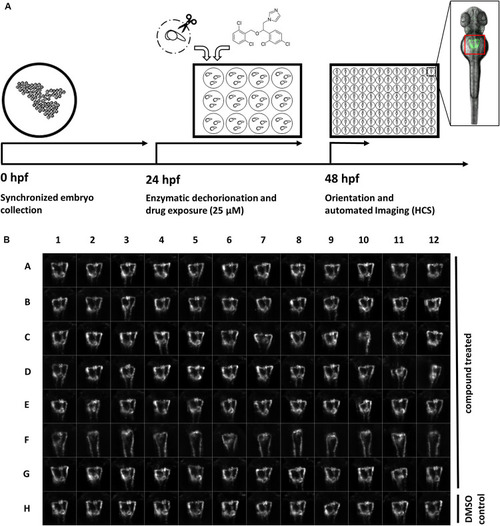
A screening workflow to score potential nephrotoxicity of approved drugs. |
|
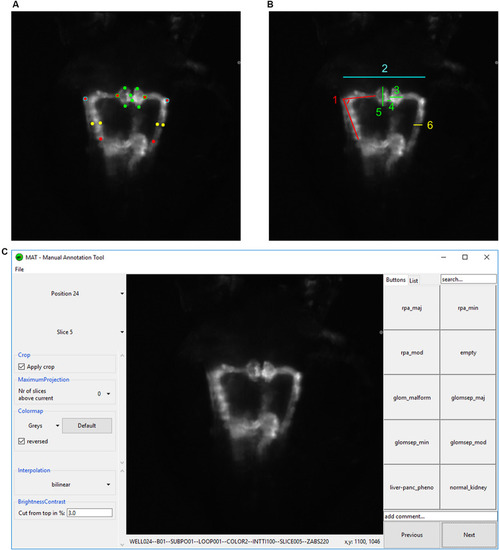
Analysis and annotation of pronephric phenotypes. |
|
Overview of scored quantitative morphometric parameters and qualitative manual annotations. |
|
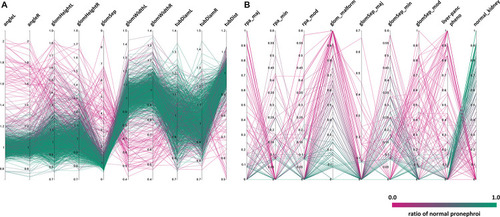
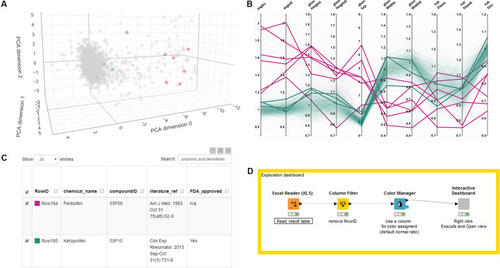
Distribution of phenotypic features upon compound treatment. Parallel coordinates plot visualizations showing the distribution of phenotypic features of pronephroi upon compound treatment. Each line plot represents a single compound treatment. In total, 1237 compound treatments are shown. The color of lines indicates the ratio of embryos within one treatment group scored as “normal_kidney” using manual annotation (from 0% (magenta, all abnormal) to 100% (green, all normal)). |
|
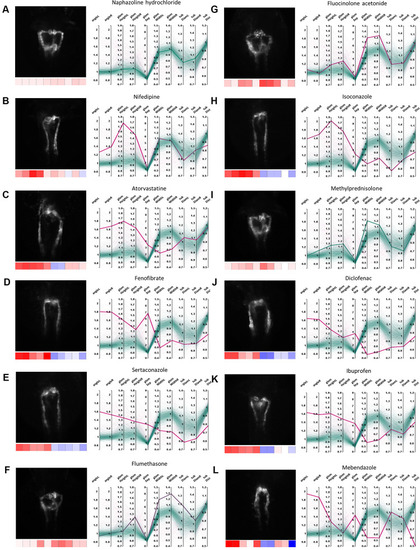
Examples of pronephric phenotypes. Illustrative examples of drug induced phenotypic changes for several compound classes. For each compound a thumbnail image, a heat map z-score visualization (below thumbnail) and a parallel coordinates plot of fold changes of quantitative morphological features are shown. In the parallel coordinates plots, thick lines indicate the shown compound and thin lines represent all other compound treatments; color codes as in |
|
Exploring and browsing the generated zebrafish embryo nephrotoxicity dataset. Shown are screenshots of an interactive exploration tool generated using KNIME (see |