- Title
-
In vivo cell biological screening identifies an endocytic capture mechanism for T-tubule formation
- Authors
- Hall, T.E., Martel, N., Ariotti, N., Xiong, Z., Lo, H.P., Ferguson, C., Rae, J., Lim, Y.W., Parton, R.G.
- Source
- Full text @ Nat. Commun.
|
EXPRESSION / LABELING:
|
|
|
|
|
|
PHENOTYPE:
|
|
|
|
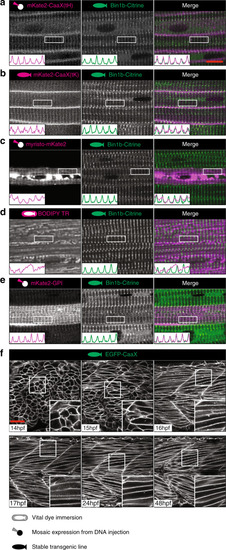
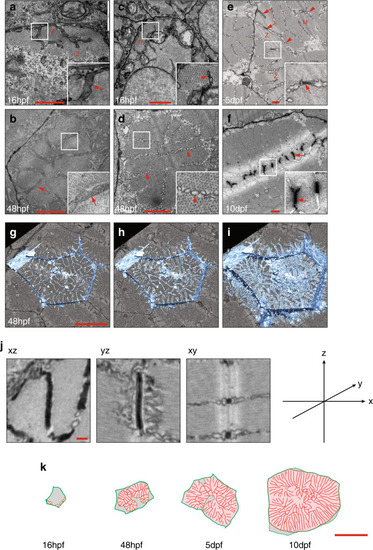
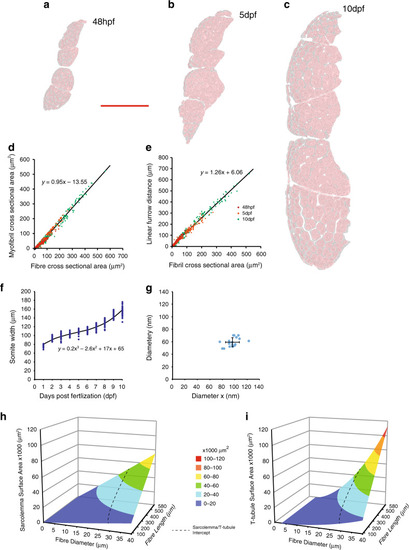
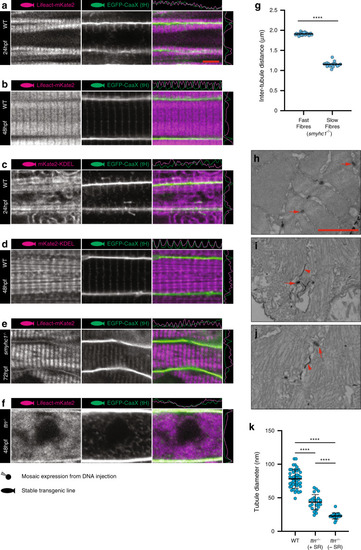
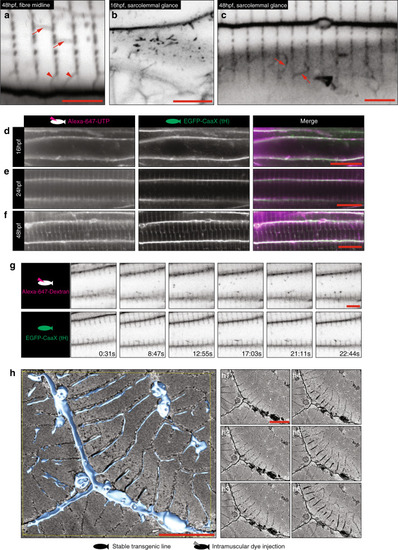
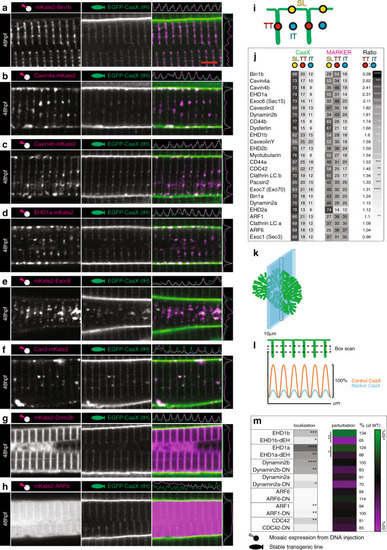
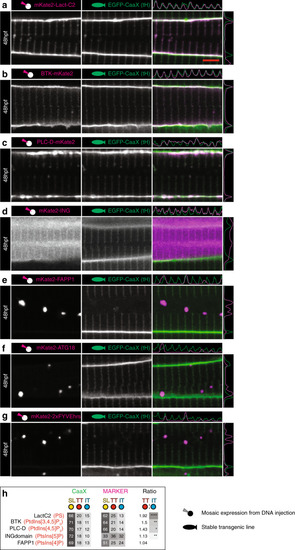
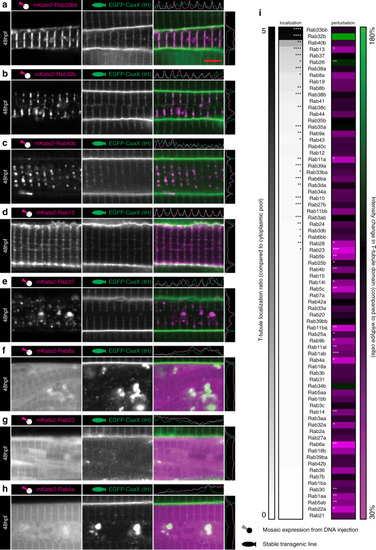
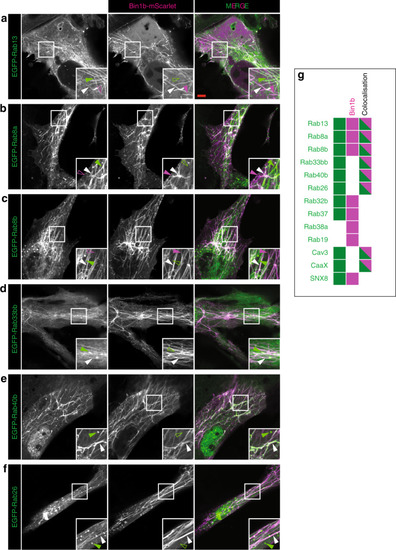
Images of the top seven markers for T-tubule localisation, compared to a non-tubule marker (ARF6). |
|
|
|
|
|
( |