- Title
-
Efficient and risk-reduced genome editing using double nicks enhanced by bacterial recombination factors in multiple species
- Authors
- He, X., Chen, W., Liu, Z., Yu, G., Chen, Y., Cai, Y.J., Sun, L., Xu, W., Zhong, L., Gao, C., Chen, J., Zhang, M., Yang, S., Yao, Y., Zhang, Z., Ma, F., Zhang, C.C., Lu, H.P., Yu, B., Cheng, T.L., Qiu, J., Sheng, Q., Zhou, H.M., Lv, Z.R., Yan, J., Zhou, Y., Qiu, Z., Cui, Z., Zhang, X., Meng, A., Sun, Q., Yang, Y.
- Source
- Full text @ Nucleic Acids Res.
|
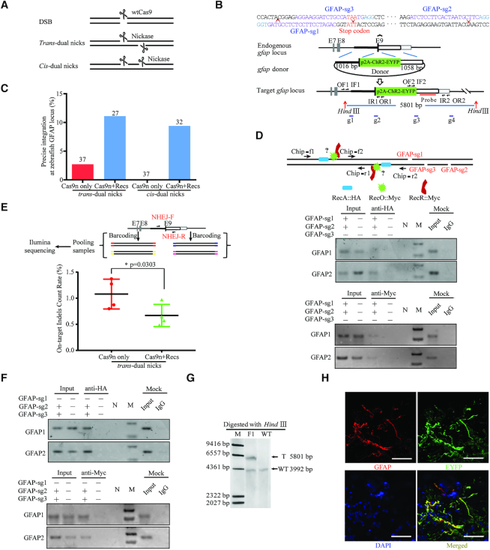
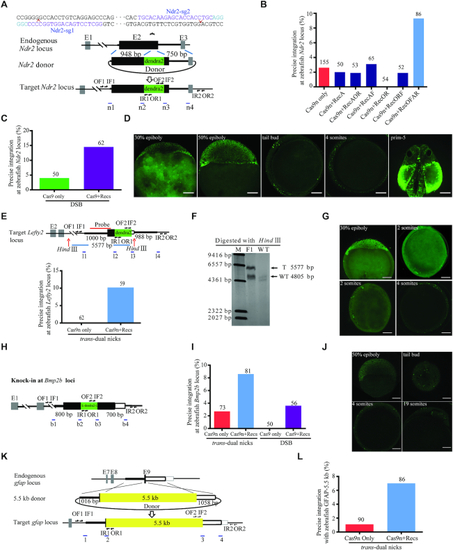
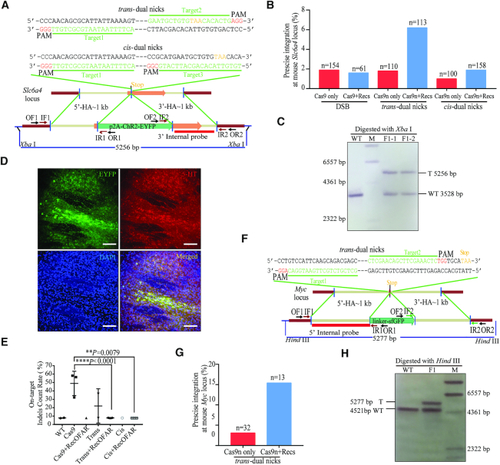
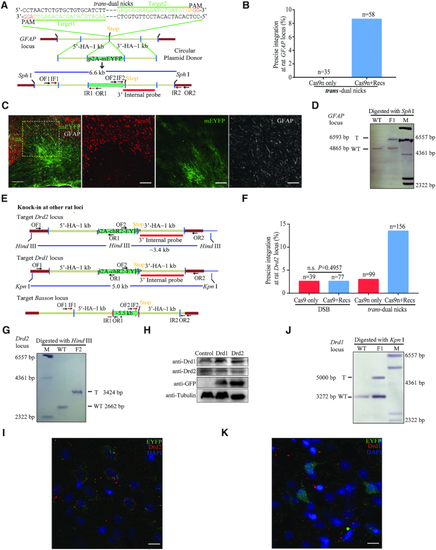
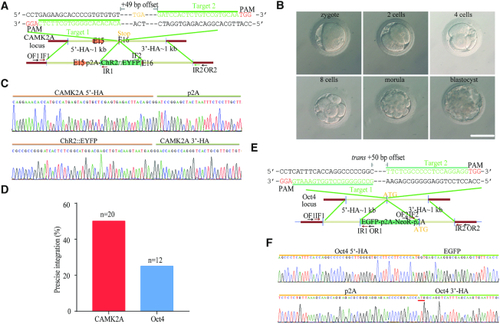
Design and optimisation of dual nicks-based gene KI at the |
|
RecOFAR facilitate dual nicks-based efficient gene KI at multiple loci in zebrafish. ( |
|
|
|
|
|
|
|
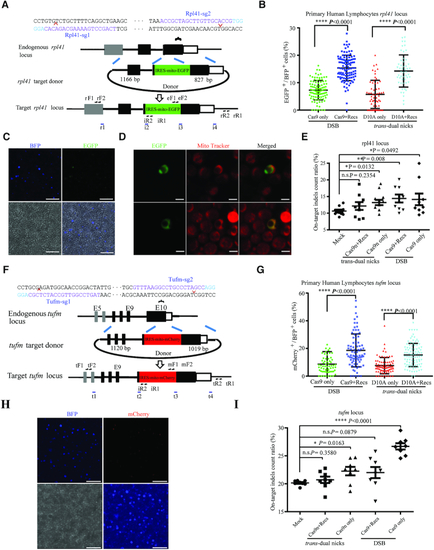
NEO achieved efficient integration in non-dividing primary human PBLCs. ( |