- Title
-
Cdh2 coordinates Myosin-II dependent internalisation of the zebrafish neural plate
- Authors
- Araya, C., Häkkinen, H.M., Carcamo, L., Cerda, M., Savy, T., Rookyard, C., Peyriéras, N., Clarke, J.D.W.
- Source
- Full text @ Sci. Rep.
|
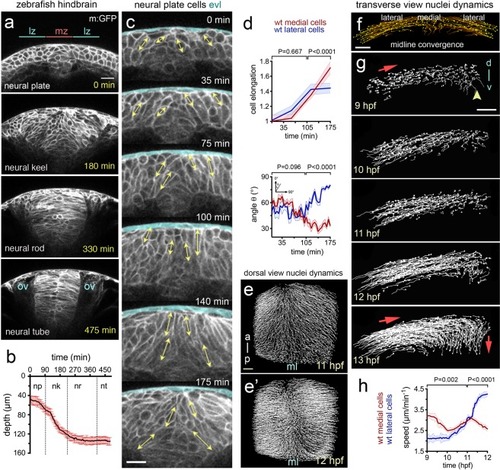
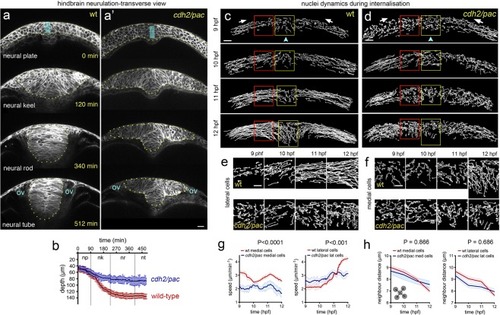
Cell and tissue dynamics during zebrafish neural plate internalisation. ( |
|
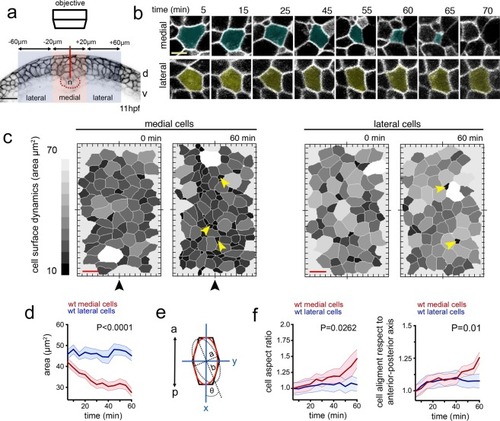
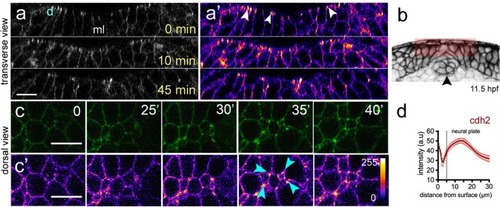
Dorsal cell surface dynamics during internalisation. ( |
|
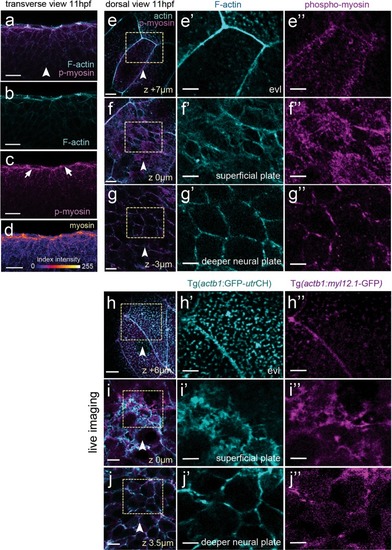
Myosin and actin in the neural plate. ( EXPRESSION / LABELING:
|
|
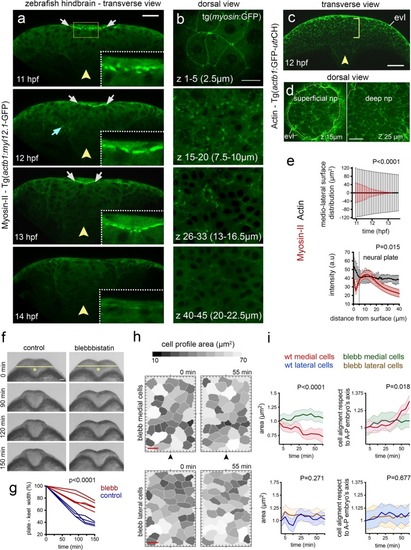
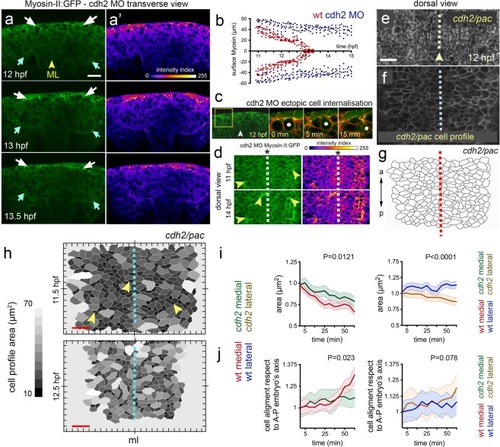
Myosin distribution and function during convergence and internalisation. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
Defective cell and tissue internalisation in PHENOTYPE:
|
|
Cdh2 distribution during neural plate internalisation. ( EXPRESSION / LABELING:
|
|
Cdh2 is required to organise midline Myosin-II during internalisation. ( EXPRESSION / LABELING:
PHENOTYPE:
|
|
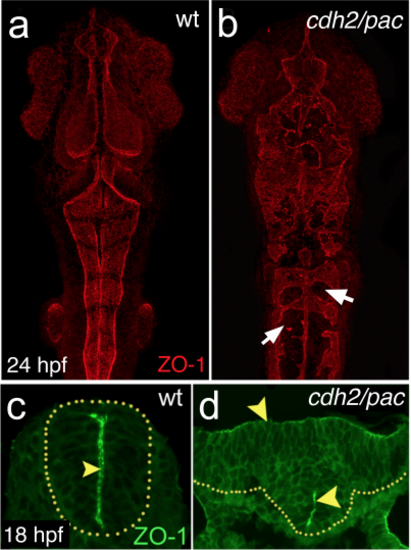
Depletion of cdh2 leads to defective neurulation and abnormal apicobasal polarity (a-b) Maximum projections of dorsal confocal pictures showing abnormal neural tube morphology and disorganised apical midline seam specialisation in cdh2/pac by 24 hpf, as revealed by apical marker, ZO-1 (arrows). Anterior is up. (c-d) Cross-section analysis in cdh2/pac mutants reveal disrupted apical midline seam organisation (arrowheads) at hindbrain levels. |
|
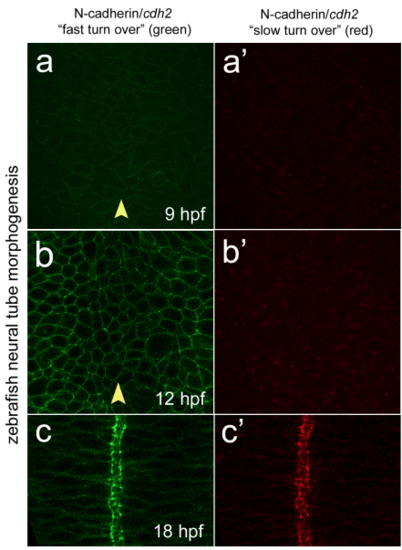
Cdh2 turnover through zebrafish neurulation (a) Dorsal single confocal picture of zebrafish neural plate at 9 hpf showing Cdh2- fast/dynamic “green” turnover in TgBAC(cdh2:cdh2-tFT) 24. (a’) Dorsal single confocal picture of zebrafish neural plate at 9 hpf showing n-cadherin slow/stable “red” turnover in TgBAC(cdh2:cdh2-tFT). (b) Dorsal confocal picture of zebrafish neural plate at 12 hpf showing Cdh2 “green” turnover. (b’) Dorsal confocal picture of zebrafish neural plate at 12 hpf showing Cdh2 “red” turnover. (c) Dorsal confocal picture of zebrafish neural tube at 18 hpf showing Cdh2 “green” turnover. (c’) Dorsal confocal picture of zebrafish neural tube at 18 hpf showing Cdh2 “red” turnover. Arrowhead indicates tissue midline |