- Title
-
miR-216a regulates snx5, a novel notch signaling pathway component, during zebrafish retinal development
- Authors
- Olena, A.F., Rao, M.B., Thatcher, E.J., Wu, S.Y., Patton, J.G.
- Source
- Full text @ Dev. Biol.
|
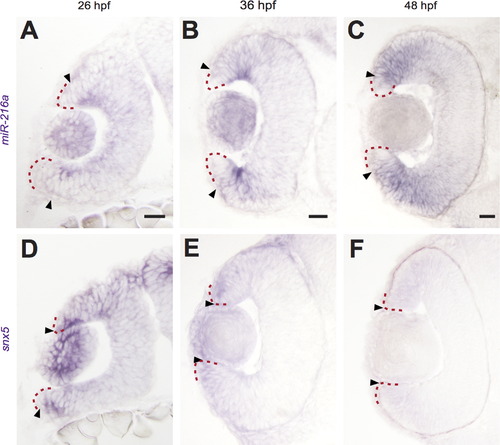
miR-216a and snx5 have complementary expression patterns during development. Transverse sections of whole mount in situ hybridizations for miR-216a and snx5 at 26 (A, D), 36 (B, E), and 48 h (C, F) post-fertilization (hpf). miR-216a expression spreads from the center of the developing retina toward the periphery. snx5 is detected in a complementary pattern becoming increasingly restricted over time to a small number of cells at the far periphery of the developing retina. Arrowheads indicate the extent of signal, the red dashed line indicates the lateral edge of the optic cup. Scale bar: 20µm. |
|
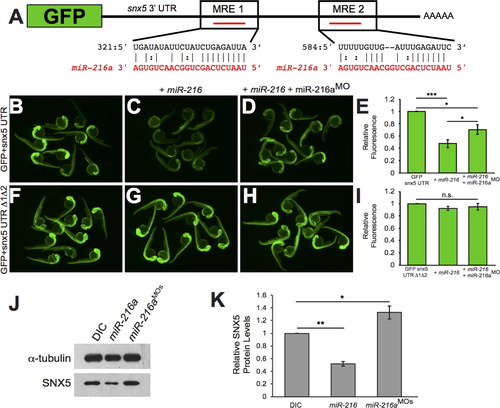
snx5 is a target of miR-216a. The coding sequence of GFP was fused to the 32UTR of snx5. Predicted pairing of each MRE in the 32 UTR (black) and miR-216a (red) are pictured. (B) Embryos injected at the 1-cell stage with GFP-snx5 32 UTR reporter mRNA alone, with miR-216a (C), or with miR-216a and miR-216aMO (D) were imaged using a fluorescence dissecting scope at 1 dpf. (F) Both MREs were deleted from the GFP-snx5 32 UTR reporter. Embryos injected at the 1-cell stage with this mRNA alone, with miR-216a (G), or with miR-216a and miR-216aMO (H) were imaged at 1 dpf using a fluorescence dissecting scope. (E, I) Relative fluorescence was quantified using ImageJ, and comparisons were made using one-way ANOVA with Bonferroniós correction. (J) Western blots for SNX5 and alpha-tubulin were performed on protein lysates from 1 dpf zebrafish injected at 1-cell stage with dye control (DIC), miR-216a, or miR-216aMOs. (K) Band density was quantified using ImageJ, and comparisons were made using one-way ANOVA with Bonferroniós correction. *, p<0.05; **, p<0.01; ***, p<0.001. Error bars show SEM. |
|
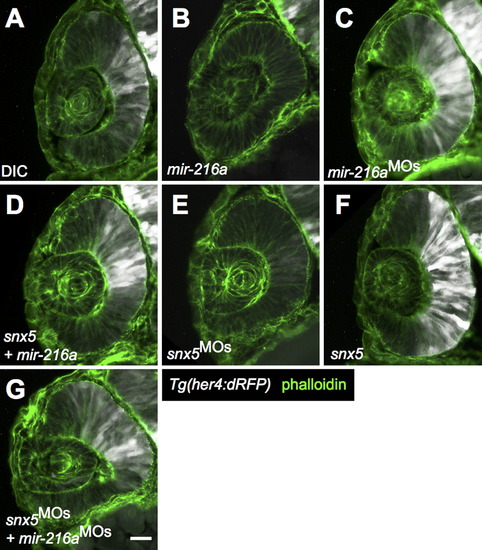
miR-216a and snx5 regulate Notch activation. Transverse sections of developing retinas from 30 h post-fertilization (hpf) Tg(her4:dRFP) embryos were injected with dye control (DIC; A), miR-216a (B), miR-216aMOs (C), snx5MOs (E), or snx5 mRNA (F). Reporter expression (white) indicates changes in the zone of Notch activation. Partial rescue of Notch activity is shown in (D) and (G) where embryos were co-injected with combinations of either snx5 and miR-216a (D) or snx5MOs and miR-216aMOs (G). Sections were stained with Alexa Fluor 488-conjugated phalloidin (green) to visualize cell boundaries. Scale bar: 20 µm. |
|
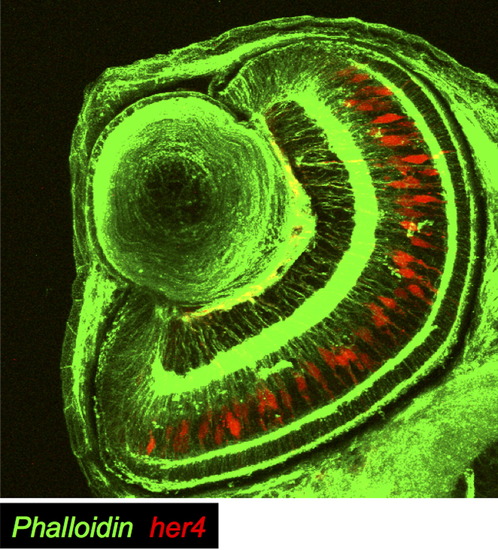
Notch activation in Müller glia at 65 hpf. In a cross section of Tg(her4:dRFP) fish at 65 hpf, Notch activation (in red) was detected primarily in Müller glia. Cell membranes are labeled with phalloidin, here visualized in green. EXPRESSION / LABELING:
|
|
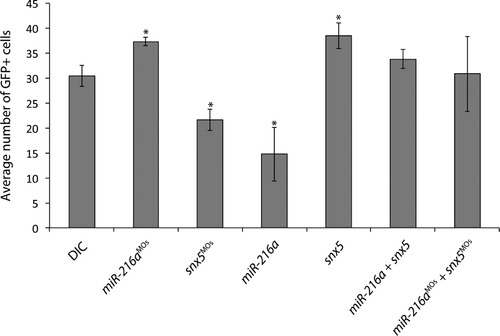
miR-216a and snx5 regulate Müller glia cell numbers. Tg(gfap:GFP) transgenic zebrafish were injected as indicated, grown to 65 hpf, and GFP+ cell numbers were counted. Compared to DICs, injection of miR-216aMOs or snx5 caused a significant increase in GFP+ cells (p<0.05). Injections with miR-216a or snx5MOs caused a significant decrease in GFP+ cells (p<0.05). Partial rescue of GFP+ cell counts was observed in embryos co-injected with combinations of either snx5 and miR-216a, or snx5MOs and miR-216aMOs. Error bars=SEM. PHENOTYPE:
|
|
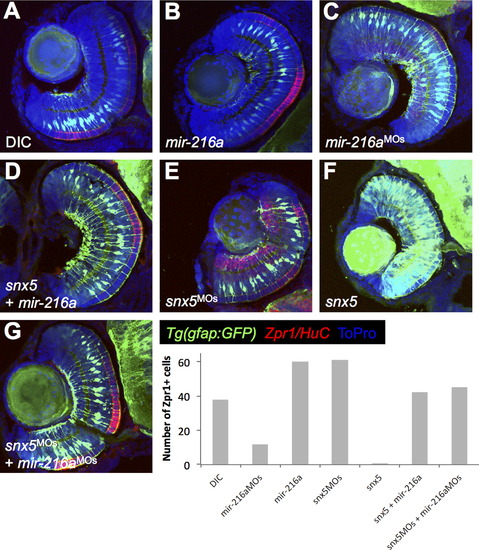
Inverse correlation between MG numbers and cone photoreceptor staining. Tg(gfap:gfp) embryos were injected with dye control (DIC; A), miR-216a (B), miR-216aMOs (C), snx5MOs (E), or snx5 mRNA (F) at the 1-cell stage, fixed at 65 hpf, and transverse sections of developing retinas were obtained. Immunohistochemistry was performed using antibodies to identify cone photoreceptors in the outer nuclear layer (Zpr-1) or amacrine/ganglion cells in the inner nuclear layer and the ganglion cell layer (HuC). Changes in Müller glia cell numbers led to consistent changes in cone photoreceptor numbers. Zpr-1 staining increased in embryos injected with mir-216a or snx5MOs and decreased in embryos injected with mir-216aMOs or snx5 compared to embryos injected with dye. Partial rescue of Zpr-1 levels is shown in (D) and (G) where embryos were co-injected with combinations of either snx5 and miR-216a (D) or snx5MOs and miR-216aMOs (G). Amacrine and ganglion cell numbers demonstrated similar, though less striking and less consistent changes compared to cone photoreceptors. Nuclei were marked by staining with To-Pro. |
|
miR-216a and EXPRESSION / LABELING:
|
|
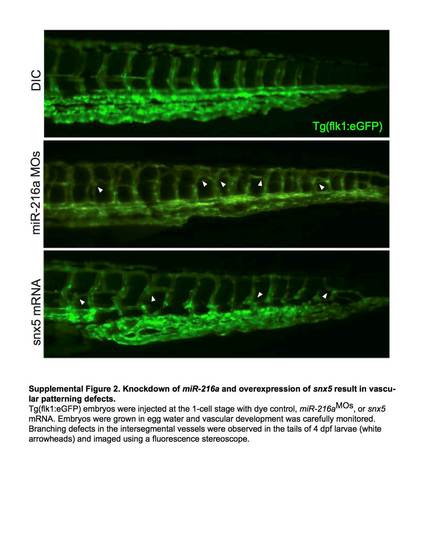
Knockdown of miR-216a and overexpression of snx5 result in vascular patterning defects. Tg(flk1:eGFP) embryos injected at the 1-cell stage with dye control, miR-216aMOs, or snx5 mRNA. Embryos were grown in egg water and vascular development was carefully monitored. Branching defects in the intersegmental vessels were observed in the tails of 4dpf larvae (white arrowheads) and imaged using a fluorescence stereoscope. |
|
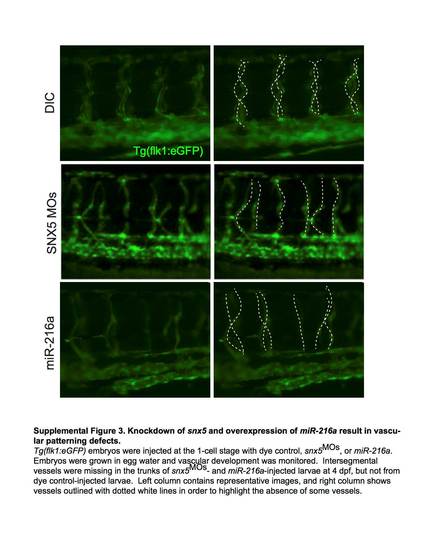
Knockdown of snx5 and overexpression of miR-216a result in vascular patterning defects. Tg(flk1:eGFP) embryos were injected at the 1-cell stage with dye control, snx5MOs, or miR-216a. Embryos were grown in egg water and vascular development was monitored. Intersegmental vessels were missing in the trunks of snx5MOs - and miR-216a-injected larvae at 4dpf, but not from dye control-injected larvae. Left column contains representative images, and right column shows vessels outlined with dotted white lines in order to highlight the absence of some vessels. |
|
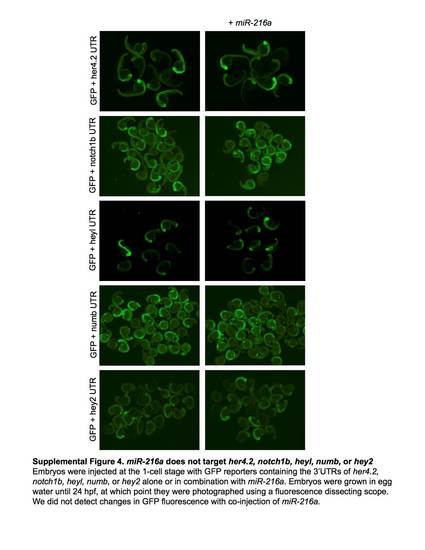
miR-216a does not target her4.2, notch1b, heyl, numb, or hey2. Embryos were injected at the 1-cell stage with GPF reporters containing the 3'UTRs of her4.2, notch1b, heyl, numb, or hey2 alone or in combination with miR-216a. Embryos grown in egg water until 24 hpf, at which point they were photographed using a fluorescence dissecting scope. We did not detect changes in GFP fluorescence with co-injection of miR-216a. |
|
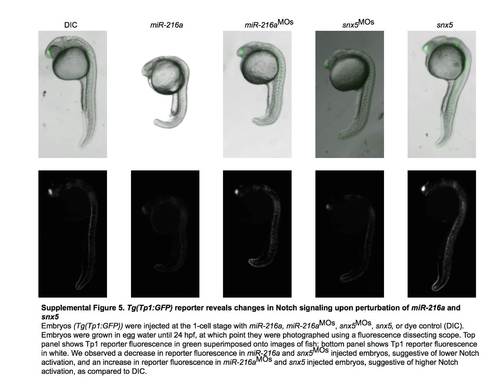
Tg(Tp1:GFP) reporter reveals changes in Notch signaling upon perturbation of miR-216a and snx5. Embryos (Tg(Tp1:GFP)) were injected at the 1-cell stage with miR-216a, miR-216aMOs, snx5MOs, snx5, or dye control (DIC). Embryos were grown in egg water until 24 hpf, at which point they were photographed using a fluorescence dissecting scope. Top panel shows Tp1 reporter fluorescence in green superimposed onto images of fish; bottom panel shows Tp1 reporter fluorescence in white. We observed a decrease in reporter fluorescence in miR-216a and snx5MOs injected embryos, suggestive of lower Notch activation, and an increase in reporter fluorescence in miR-261aMOs and snx5 injected embryos, suggestive of higher Notch activation, as compared to DIC. |
|
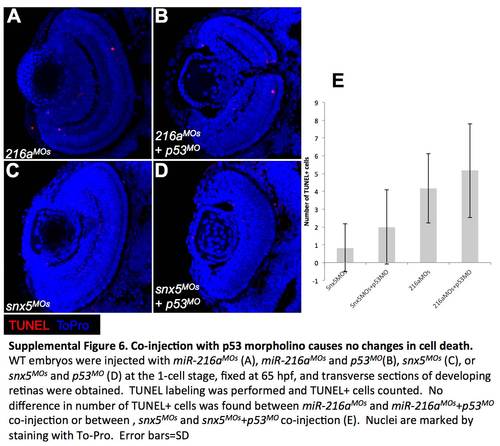
Co-injection with p53 morpholino causes no changes in cell death. WT embryos were injected with miR-216aMOs (A), miR-216aMOs and p53MO (B), snx5MOs (C), or snx5MOs and p53MO (D) at the 1-cell stage, fixed at 65 hpf, and transverse sections of developing retinas were obtained. TUNEL labeling was performed and TUNEL+ cells counted. No difference in number of TUNEL+ cells was found between miR-216aMOs and miR-216aMOs+p53MO co-injection or between, snx5MOs and snx5MOs+p53MO co-injection (E). Nuclei are marked by staining with To-Pro. Error bars = SD. |
Reprinted from Developmental Biology, 400(1), Olena, A.F., Rao, M.B., Thatcher, E.J., Wu, S.Y., Patton, J.G., miR-216a regulates snx5, a novel notch signaling pathway component, during zebrafish retinal development, 72-81, Copyright (2015) with permission from Elsevier. Full text @ Dev. Biol.