- Title
-
Molecular and Cellular Analysis of the Repair of Zebrafish Optic Tectum Meninges Following Laser Injury
- Authors
- Banerjee, P., Joly, P., Jouneau, L., Jaszczyszyn, Y., Bourge, M., Affaticati, P., Levraud, J.P., Boudinot, P., Joly, J.S.
- Source
- Full text @ Cells
|
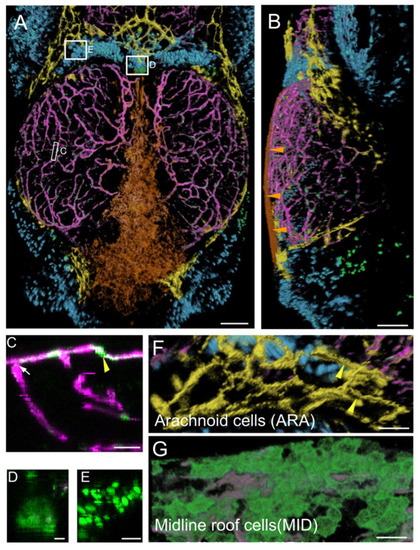
Main expression domains of the PDGFRβ-gal4:UAS-EGFP line as seen by high-content confocal imaging of 21 dpf juveniles. Three main meningeal domains were segmented and color-coded in orange, yellow and blue. (A,B). 3D reconstructions using the AMIRA software. Midline roof cells (MID): orange; arachnoid cells (ARA): yellow; red blood cells (RBC): blue; vessels: magenta. (A). Dorsal view. White rectangles indicate locations of details in (C–E). (B). Lateral view. Orange arrowheads show the flat MID. (C). XZ thick optical section showing one pericyte (yellow arrowhead) and one vessel (white arrow). (D). Dorsal view of choroid plexus. (E). Red Blood Cells (RBC). (F). Dorsal view of ARA showing typical elongated and square shapes (as described for mammalian interstitial arachnoid cells) and granulations (yellow arrowheads). (G). Dorsal view of MID stained with Aqp1a.1 antibody showing their round and flat ameboid-like shape. Scale bars: (A,B): 100 μm. (C–G): 50 μm. (D–E): 10 μm. |
|
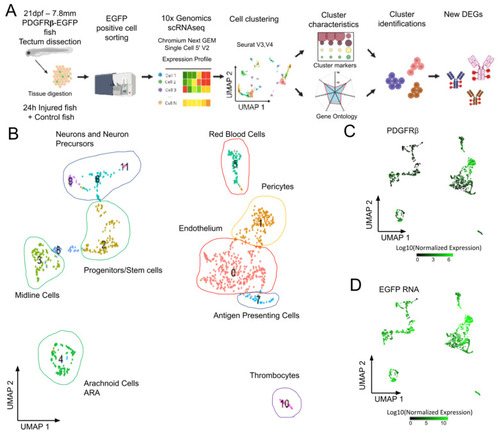
Analysis of scRNAseq data highlights cell heterogeneity of GFP positive cells from dissected tecta of PDGFRβ-gal4:UAS-EGFP juveniles. (A). Experimental workflow for the collection and isolation of OT cells. (B). Uniform manifold approximation and projection (UMAP) showing cell populations from control 21 dpf juveniles. The projection was performed with the 30 first principal component analysis of gene expression. Each point represents a single cell with a colour indicating its membership to a cluster. 0: KDRL- Endothelium; 1: NDUFA.4L2A-Pericyte; 2: RPS2-Progenitors, Stem cells; 3: ZIC4- Midline Cells; 4: AQP1A.1-Arachnoid Cells; 5: HBAA1-Red Blood Cells; 6: ELAVL4- Neurogenic Precursors; 7: CD74A-Antigen Presenting Cells; 8: Uncharacterized; 9: NEUROD1- Neurons and Precursors; 10: THBS1B- Thrombocytes; 11: HPCA- Neurons. (C). Featureplot highlighting PDGFRβ transcript levels in cells. (D). Featureplot of GFP transcript levels. PDGFRβ expression is correlated to GFP showing that most GFP+ cells express PDGFRβ. |
|
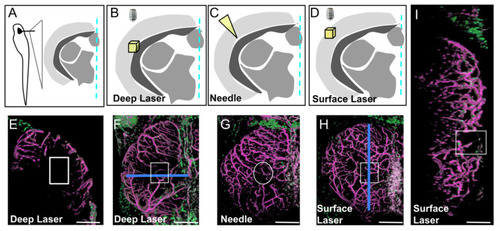
Three types of injury of the optic tectum (OT) at 21 dpf. (A). Schematic cross section at the level of the center of the OT with approximate location of details in (B–D). (B,E,F). Deep biphoton laser injury. (C,G). Needle injury. (D,H,I). Surface biphoton laser injury. Blue lines in (F,H) indicate locations of sections shown in (E) and (I). (E,I). Cross and parasagittal sections showing locations of wounds (white rectangles). (F–H). Dorsal views with location of injuries (white squares and circle). Anterior at the top. Midline on the right. Scale bars: (E–I): 100 μm. |
|
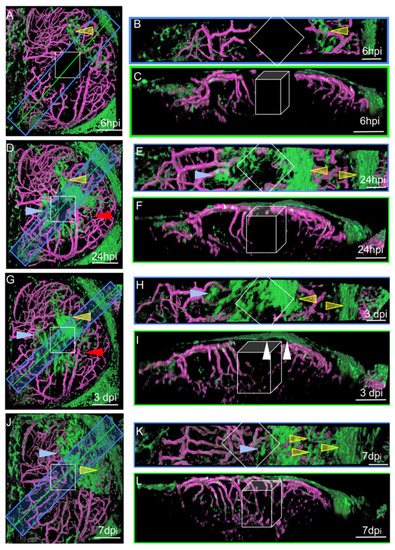
Surface laser injury of the optic tectum (OT) at 21 dpf. (A,D,G,J). Dorsal views. Blue rectangles: locations of horizontal sections in the right panels. Yellow lines: locations of XZ sections in the right panels. Anterior at the top. Midline on the left. (B,E,H,K) Horizontal thick sections. (C,F,I,L) XZ sections. (I). At 3 dpi, multiple layers of arachnoid cells are visible over the wound (white arrowheads). Putative injured zones: white squares and boxes. ARA-like cells: yellow arrowheads. MID-like cells: blue arrowheads. Fibroblast-like cells: red arrows. Scale bars: (A–L): 100 μm. (B,E,H,K): 50 μm. |
|
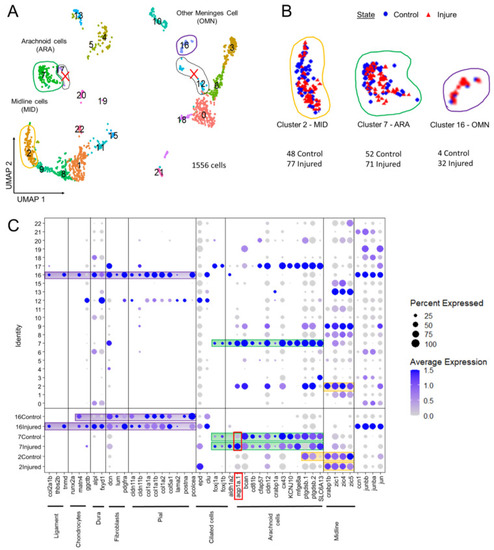
Analysis of scRNAseq data highlight cell heterogeneity of GFP cells from dissected tectum of PDGFRβ-gal4;UAS: EGFP in 21 dpf injured fish and 21 dpf control fish. (A). Uniform manifold approximation and projection (UMAP) showing cell populations from 21 dpf control and injured fish. The projection was computed from the twenty first principal component analyses of gene expressions. Each point represents a single cell with a colour indicating its membership to a cluster. (B). UMAP of injured and control cells colored according to their sample types (blue circle for control fish and red triangle for injured fish). Approximately equal numbers of cells of each origin was observed in most clusters. This avoided batch effect. (C). Cluster analysis of characteristic DEGs from different cell populations including arachnoid cells (ARA) found in cluster 7, midline-like cells (MID) found in cluster 2 and other meninx cells (OMN) found in cluster 16. Cluster 17 et 12 contained DEGs similar to clusters 2, 7, 16 but these clusters were discarded from our analysis due to their small size and heterogeneity. |
|
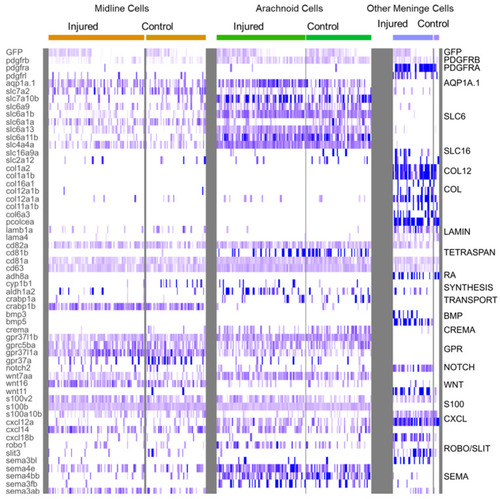
Heatmap of characteristic gene expression levels in MID, ARA and OMN cells from control and injured datasets. Each column represents a cell. Level of expression is encoded by color gradient. Strong expression is in dark blue and no expression is in white. Gene families are represented to the right of the panel in regards to the corresponding DEGs. |
|
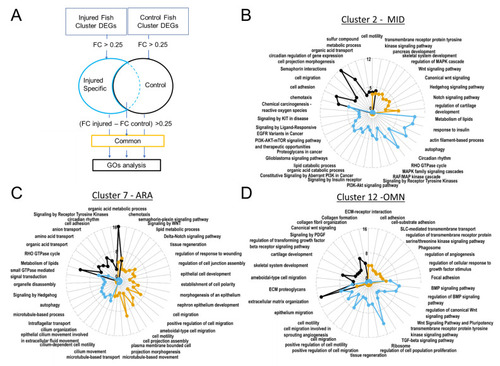
Gene ontology revealed upregulated biological processes. In ARA gene lists corresponding to genes specifically present in samples from injured fish, cilium-dependent cell motility GO and other cilium associated GOs are enriched. (A). Pipeline for filtering DEGs for GO analyses with Metascape. Radarplots of enriched Gene ontologies. Curves colored following types of filtering as explained in (A). Axis concern Zscore of GOs. Increased distance from the centre means higher significance. (B). OMN. (C). MID. (D). ARA. |
|
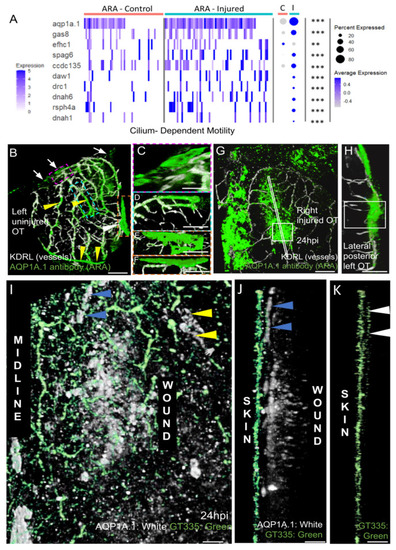
AQP1a.1 ciliated ARA migrates towards the wound. (A) Expression of AQP1a.1 and cilia motility genes in cells depending on their status show overexpression of these genes after wounds. Genes expressed <10% of cells are not presented in dotplot. Adjusted p value (** <0.01 *** <0.001) concern markers in injured status. (B) Dorsal view on an uninjured OT with ARA (white arrows), MID (white arrowhead), and vessels (yellow arrowheads) stained by the AQP1a.1 antibody. Anterior at the top. Midline on the left. (C) Detailed dorsal view of ARA. (D) Detailed side view of ARA. (E) Detailed dorsal view of MID. (F) Detailed side view of MID. (G) At one dpi, strongly positive AQP1a.1 cells are visible over the wound (white arrowheads). (H) Strong and thick AQP1a.1 staining over the wound. (I) AQP1a.1-positive cells over and around the wound. Peripheral ARA-like cells: yellow arrowheads. (J) Migrating ARA stained by AQP1a.1: blue arrowheads. Lateral ARA: yellow arrowheads. (K) Migrating AQP1a.1-positive ARA are ciliated as shown by the GT335 staining: white arrowheads. AQP1a.1-positive cells do not exhibit any cilium/GT335 staining. Scale bars: (A,F): 100 μm; (B–E,G): 50 μm; (H–J): 10 μm. |