- Title
-
Potassium Channel-Associated Bioelectricity of the Dermomyotome Determines Fin Patterning in Zebrafish
- Authors
- Silic, M.R., Wu, Q., Kim, B.H., Golling, G., Chen, K.H., Freitas, R., Chubykin, A.A., Mittal, S.K., Zhang, G.
- Source
- Full text @ Genetics
|
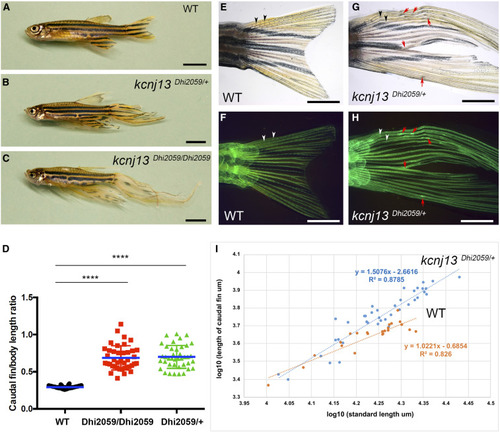
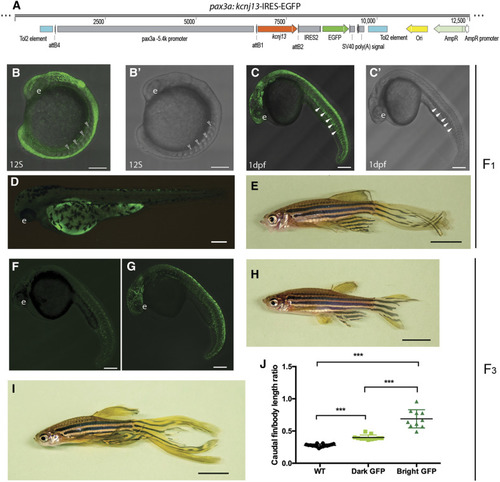
Fins are elongated in the adult zebrafish mutant Dhi2059. (A) WT fish with normal-sized fins. (B and C) Both one- and two-copy transallelic mutants of Dhi2059 possessed elongated paired and median fins. Bars, 50 mm in (A–C). (D) The lengths of caudal fins in Dhi2059 mutants are significantly longer compared to WT siblings. Fish were generated by PHENOTYPE:
|
|
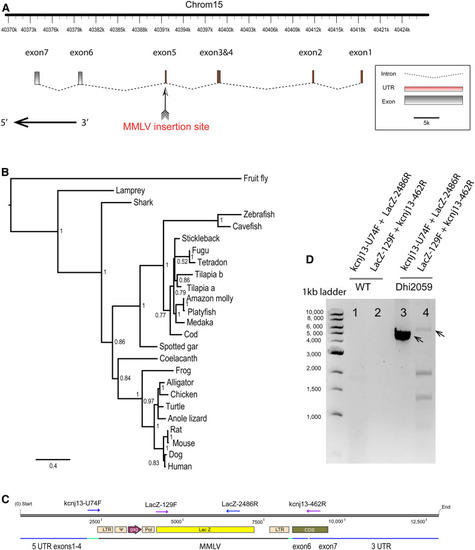
Viral insertion was identified in the |
|
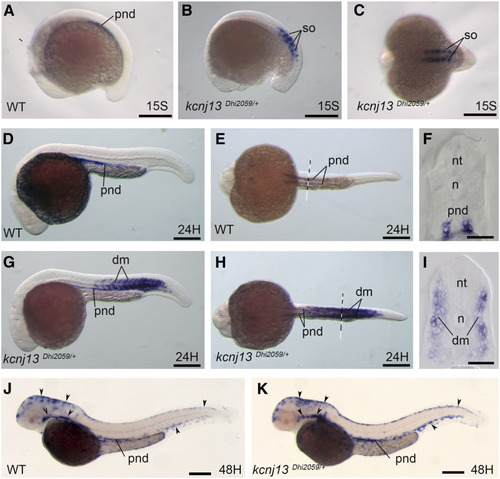
EXPRESSION / LABELING:
PHENOTYPE:
|
|
Dhi2059 long-finned phenotype was only able to be rescued by a PHENOTYPE:
|
|
Transient ectopic expression of EXPRESSION / LABELING:
|
|
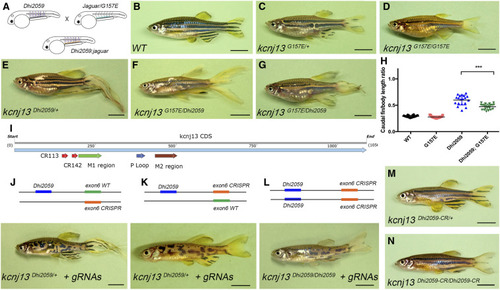
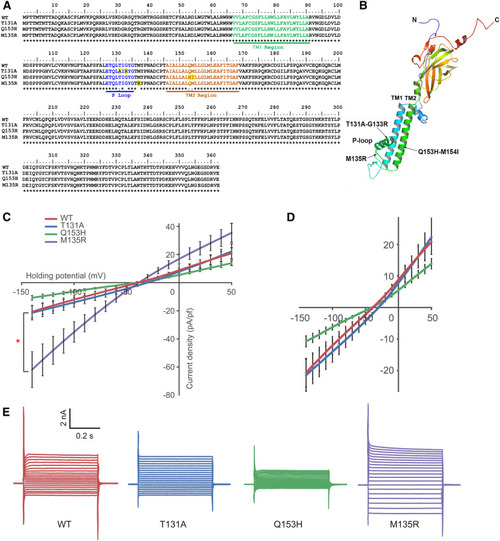
Generation of Kcnj13 mutants with different potassium conductance. (A) Protein sequence alignment of site-directed mutants. TM1, TM2, and P-loop domains are highlighted in green, brown, and blue, respectively. Altered amino acids are highlighted in yellow. (B) A computation-predicted three-dimensional structure of the Kcnj13 subunit. Mutated sites are indicated with arrows. (C) Conductance characterization of three Kcnj13 mutants compared to WT showing decreased conductance of Q153H ( |
|
Long-finned phenotype is dependent on potassium conductance. (A–E) Overexpression of |
|
Transient ectopic expression of multiple potassium channel genes also induces long-finned phenotype. (A) WT adult control fish without any injection. (B) Representative injected adult F0 fish image of human |
|
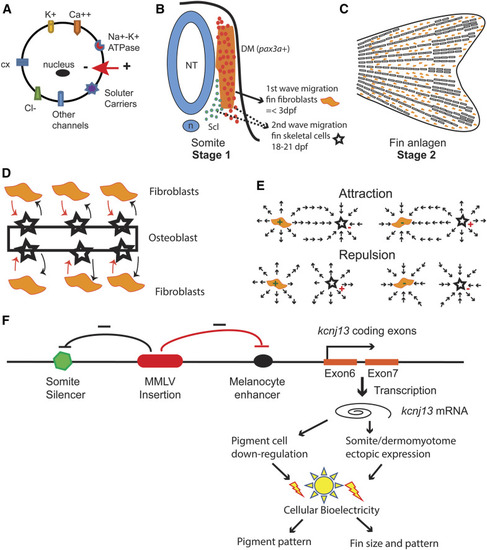
Model of |