- Title
-
High-resolution analysis of CNS expression patterns in zebrafish Gal4 enhancer-trap lines
- Authors
- Otsuna, H., Hutcheson, D., Duncan, R., McPherson, A., Scoresby, A., Gaynes, B., Tong, Z., Fujimoto, E., Kwan, K., Chien, C., Dorsky, R.
- Source
- Full text @ Dev. Dyn.
|
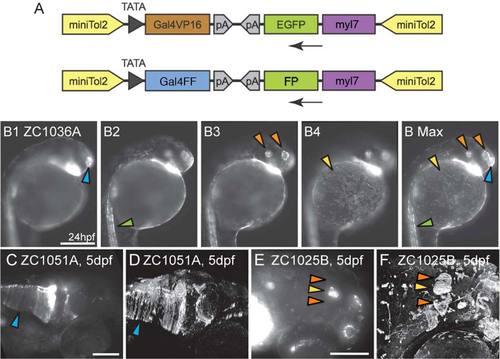
Gal4 enhancer trap constructs and imaging. A: Enhancer trap constructs used to generate all lines. B1-B Max: To convey as much information as possible, compound microscopic images were taken at multiple depths (B1–B4) and used to generate Maximum Intensity Projections (B Max) in which signal from multiple focal depths is visible. Arrowheads mark the olfactory placode (blue), spinal neurons (green), otic vesicle and retina (orange) and epithelial cells (yellow). Images B1–B Max are lateral views. C–F: Confocal images enhance fine details relative to compound microscopic images. Glial endfeet are clearly visible in the confocal image (D, Arrowhead) but are not clear in the compound scope image (C, Arrowhead). Habenulae are visible in confocal (F, Orange arrowheads) but not compound (E, Orange arrowheads) image. (E,F) Pineal gland marked by yellow arrowhead. Images C–F are dorsolateral views. Scale bars = 250 µm in B1–B MAX; 250 µm in C,D; 100 µm in E,F. |
|
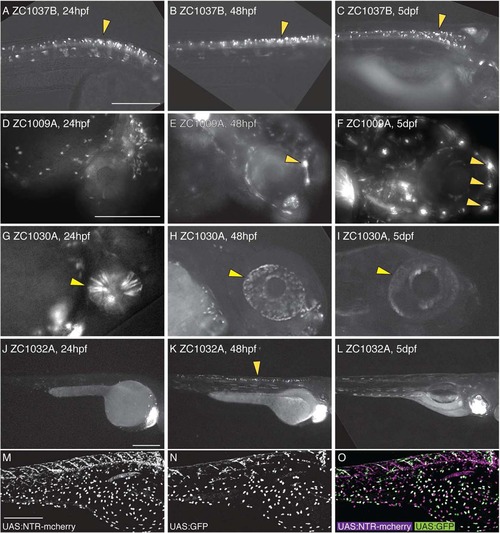
Dynamic temporal expression patterns in enhancer trap screen. Compound images taken at 24 hpf (A,D,G,J), 48 hpf (B,E,H,K), or 5 dpf (C,F,I,L) show examples of neural expression that is maintained at all ages (A–C), is activated over time (D–F), is inactivated over time (G–I), or is only present at one timepoint (J–L). Arrowheads mark spinal neurons (A–C, K), lateral line neuromasts (E,F), and the eye (G–I). UAS reporters exhibit transgene-specific variegation, leading to partially overlapping but distinct expression patterns in a single animal (M–O). All images are lateral views. Scale bar = 250 µm for A–C; 250 µm in D–I, 250 µm in J–L; 300 µm in M–O. |
|
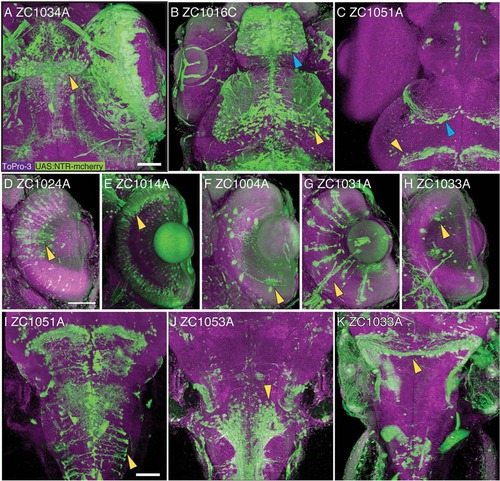
Confocal 3-D reconstruction images of 5 dpf embryos. Rostral CNS expression patterns are visible in the habenula (A, arrowhead), the telencephalon (B, blue arrowhead) and ventral tectum (B, yellow arrowhead), glial cells in the tectum (C, blue arrowhead) and midbrain–hindbrain boundary (C, yellow arrowhead). Expression is visible in major classes of retinal neurons including bipolar cells (D), photoreceptors (E), amacrine cells (F), Mueller glia (G), and ganglion cells (H). In the brainstem, expression is visible in radial glia (I), ventral neurons (J), and the rostral cerebellum (K). All images show UAS reporter (green), ToPro3 nuclear stain (magenta), and show dorsal view, with D–H showing a section through the retina. Scale bar = 50 µm. EXPRESSION / LABELING:
|
|
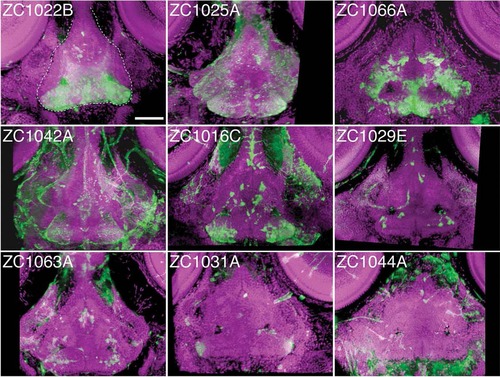
Individual enhancer traps drive various hypothalamic expression patterns. The 3D reconstructed confocal images, dorsal view, of 5 dpf whole-mount embryos stained for UAS reporter (green) and counterstained with ToPro-3 for nuclei (magenta). Partial 3D reconstruction of these images has been limited to ventral confocal planes to highlight hypothalamic expression while removing more dorsal expressing cells. Dashed white line in panel zc1022B outlines the hypothalamus. Scale bar = 50 µm. EXPRESSION / LABELING:
|
|
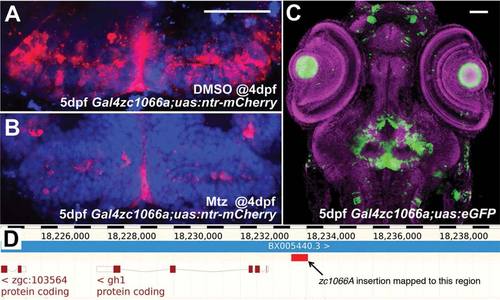
Use and characterization of a single enhancer trap line. A: Line zc1066a drives expression of NTR-mCherry in hypothalamic radial glia. B: Following incubation in metronidazole for 24 h, most mCherry+ cells are ablated and only debris remains. C: A confocal projection through the entire head of a zc1066a embryo at 5 dpf shows the specificity of this enhancer trap expression pattern. D: Using inverse-PCR, we have mapped the insertion to a genomic region immediately upstream of the gh1 mRNA. Scale bars = 50 µm. |
|
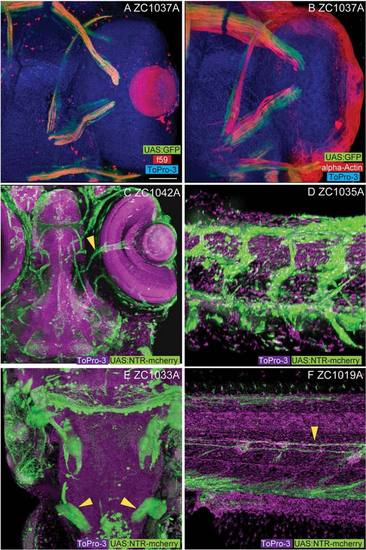
Non-CNS enhancer trap expression patterns. Among the non-CNS tissues represented in our screen, line zc1037A shows expression in slow (A, f59 positive) but not fast (B, alpha-actin) extraocular myofibers. Vascular expression is visible in both the head (C, arrowhead marks central retinal vessels, zc1042A) and body (D, zc1035A). E: Labeling of the developing pronephros (arrowheads) is visible in line zc1033A. F: Labeling of lateral line glial fibers (arrowhead) is visible in line zc1019A. Images A, B, C, and E show dorsal views. Images D and F show lateral views. Scale bar = 50 µm in A–F. EXPRESSION / LABELING:
|