- Title
-
Efficient disruption of Zebrafish genes using a Gal4-containing gene trap
- Authors
- Balciuniene, J., Nagelberg, D., Walsh, K., Camerota, D., Georlette, D., Biemar, F., Bellipanni, G., and Balciunas, D.
- Source
- Full text @ BMC Genomics
|
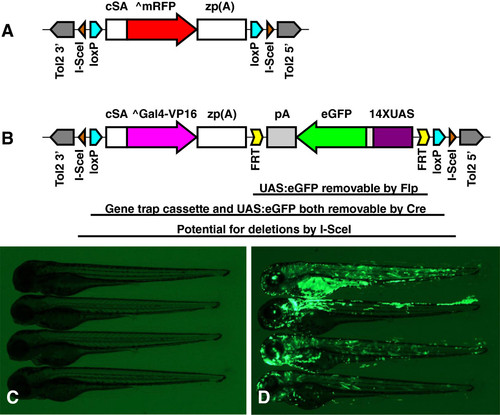
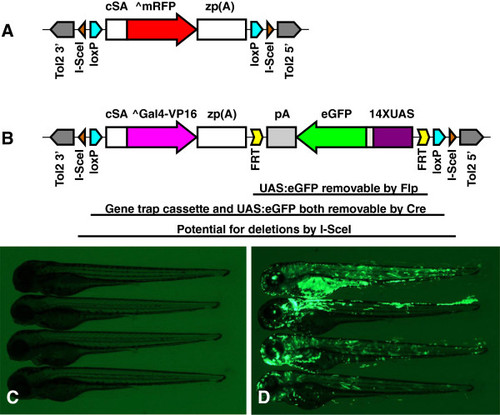
Design of GBT-B1 trap. (A) Parental vector GBT-R15 <27, 28]. cSA denotes carp β-actin splice acceptor, ^mRFP denotes AUG-less mRFP, and zp(A) denotes zebrafish β-actin 3′ untranslated region and transcriptional termination / polyadenylation cassette. Tol2 5′ and Tol2 3′ are miniTol2 transposon arms as described in [37]. (B) Features of GBT-B1 trap. ^Gal4-VP16 denotes AUG-less Gal4-VP16, and the 14XUAS:eGFP cassette is from [34]. (C) Embryos injected with GBT-B1 without Tol2 transposase mRNA. (D) Embryos injected with GBT-B1 and Tol2 transposase mRNA. |
|
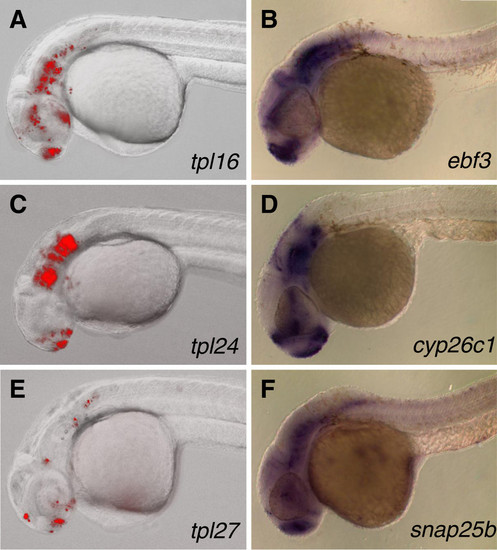
Comparison between patterns of UAS-driven mRFP fluorescence and endogenous gene expression by whole mount in situ hybridization. Embryos containing ebf3tpl16, cyp26c1tpl24 and snap25btpl27 gene traps and Tg(UAS:mRFP)tpl2 reporter where photographed on Zeiss AxioImager microscope at 30–32 hpf (A, C, and E respectively). Expression of endogenous genes detected by whole mount in situ hybridization using antisense probes against ebf3(B), cyp26c1(D) and snap25b(F) on 30 hpf embryos. EXPRESSION / LABELING:
|
|
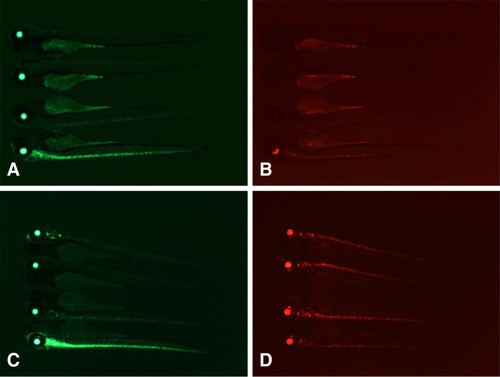
Manipulation of the nsfatpl6 gene trap using Cre and Flp recombinases. Embryos containing the nsfatpl6 gene trap and UAS:mRFPtpl2 were injected with 25 pg of Cre (A, B) or 600 pg of eFlp (C, D) mRNA. Groups of four representative embryos, with the bottom embryo representing low recombinase activity. Note simultaneous loss of GFP and RFP in panels A and B (injection of Cre RNA), and loss of GFP without loss of RFP in panels C and D (injection of eFlp RNA). |
|
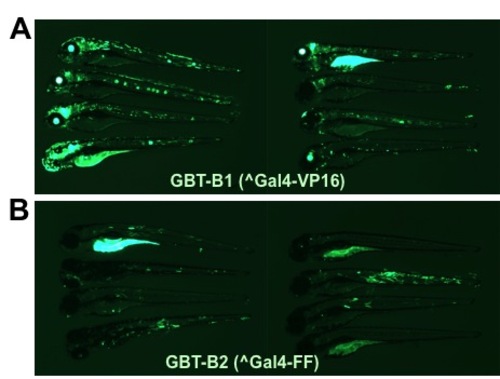
Comparison between embryos injected with GBT-B1 and GBT-B2 gene traps containing ^Gal4-VP16 and ^Gal4-FF respectively. Embryos were injected with Tol2 transposase mRNA and GBT-B1 (A) or GBT-B2 (B) plasmid DNA. At 3 dpf, random GFP-positive embryos were photographed under identical settings and images were processed identically. |

Expression of nsfa in the pancreas. Comparison between GFP expression of nsfatpl6 gene trap (A, C) and nsfa expression by whole mount in situ hybridization (B, D) in 1 dpf zebrafish embryos. Red arrow points to pancreas. |
|
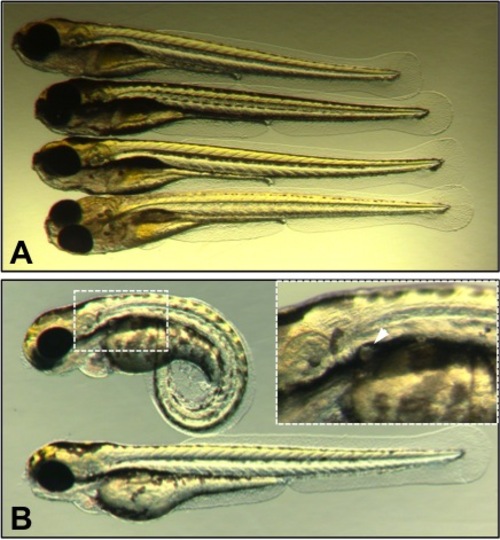
Phenotypes of nsftpl6 and flrtpl19 homozygotes. A. Larvae homozygous for nsftpl6 fail to inflate swim bladders by 6 dpf and display greatly reduced sensitivity to touch (Additional file 5: Movie 1). B. Comparison between a flrtpl19 homozygote and a wild type sibling at 3 dpf. Insert displays greater magnification of the kidney area with a cyst (white arrow). |
|
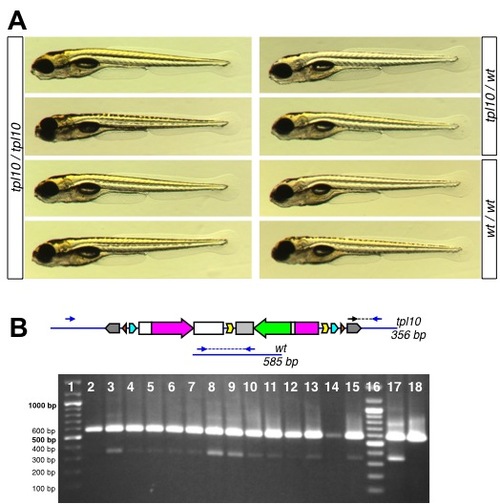
Late larval lethality of atp1a3atpl10 homozygotes. A. 5 dpf larvae homozygous for atp1a3atpl10 do not display overt embryonic phenotypes compared to heterozygous and wild type embryos. B. Genotyping of adult fish raised from atp1a3atpl10 incross embryos selected for GFP fluorescence. Diagrams on the top represent the tpl10 gene trap allele and wild type allele with expected sizes of PCR bands indicated. Genomic primers flanking transposon integration (Atp1a3a.9A1c.F and Atp1a3a.9A1A.R) are depicted as blue arrows, transposon-specific primer Tol2-R5 is depicted as a black arrow. PCR bands are shown as blue (wild type) and black (gene trap allele) dashed lines. Short PCR extension time does not allow amplification across GBT-B1. Below the diagram is a picture of a genotyping gel. Lanes 1 an 16, DNA ladder (Thermofisher Fermentas Cat #SM0331, sizes of relevant bands indicated to the left). Lanes 2–15 are PCR reactions on DNA from individual tailclips. Lanes 17 and 18 are PCR reactions performed on DNA from pools of GFP-positive (lane 17) and GFP-negative (lane 18) embryos. |
|
|
|
|
|
|