- Title
-
The ubiquitin proteasome system is required for cell proliferation of the lens epithelium and for differentiation of lens fiber cells in zebrafish
- Authors
- Imai, F., Yoshizawa, A., Fujimori-Tonou, N., Kawakami, K., and Masai, I.
- Source
- Full text @ Development
|
Zebrafish vov mutant shows defects in lens fiber cell differentiation. (A) Wild-type (wt) and vov mutant 72 hpf embryos. The vov mutant embryos exhibit hemorrhaging on the roof of the mid-hindbrain (arrowhead). (B,C) Wild-type (B) and vov mutant (C) eyes at 72 hpf. (D,E) Sections of 72 hpf wild-type (D) and vov mutant (E) eyes. Bracket indicates apoptosis near the retinal ciliary marginal zone (CMZ) (E). (F,G) Wild-type (F) and vov mutant (G) lens. Nuclei are flat and positioned along the posterior edge of the wild-type lens sphere (arrowheads, F), whereas nuclei are swollen and located in the anterior region surrounding a small lens fiber core in the vov mutant (arrowheads, G). (H,I) Expression of Pax6 (green) and ZO-1 (red) in 72 hpf wild-type (H) and vov mutant (I) lens. Arrowheads indicate aggregated ZO-1 signals in the posterior lens region of the vov mutant. (J,K) Expression of Pcna (green) and Prox1 (red) in 72 hpf wild-type (J) and vov mutant (K) lens. Arrowheads indicate the Pcna and Prox1 double-positive cells. Arrows indicate Pcna-positive cells located in the posterior edge of the lens sphere (K), most of which weakly express Prox1 (see Fig. S3D-D4 in the supplementary material). (L,M) Double labeling of 72 hpf wild-type (L) and vov mutant (M) lens with BrdU (green) and anti-pH3 antibody (red). Nuclei are stained with TOPRO3 (gray). (N,O) Expression of AQP0 (red) in 72 hpf wild-type (N) and vov mutant (O) lens. Nuclei are stained with DAPI (green). Arrowheads indicate nuclei of AQP0-positive lens fiber cells in the vov mutant, which are located in the anterior lens region (O). (P,Q) Expression of Lamin B1 in 72 hpf wild-type (P) and vov mutant (Q) lens. Arrowheads indicate wild-type lens fiber cell nuclei. White dashed lines indicate the outline of the lens sphere. (R) Schematic of wild-type and vov mutant lens. Lens epithelium (ep) expresses Pax6 (green) and its marginal cells are associated with retinal CMZ. After passing over the equator, lens epithelial cells transiently express Prox1 (light green) and differentiate into lens fiber cells (fib). Newly differentiating lens fiber cells (light blue) express AQP0 (orange) and their nuclei become flat and are subsequently denucleated. In the vov mutant, cell proliferation of lens epithelium is decreased and lens fiber cells are disorganized in morphology. ZO-1 signals are indicated by red circles. Anterior is left and dorsal is up in all the panels showing the lens (except in Fig. 3E,F and Fig. 6). Scale bars: 50 µm, except 250 µm in A. EXPRESSION / LABELING:
PHENOTYPE:
|
|
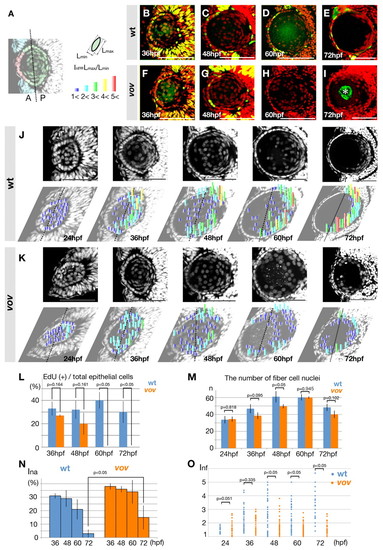
The shape, position and nuclei number of lens fiber nuclei are affected in the vov mutant. (A) Color key for lens epithelial cells and lens fiber cells. From 36 to 72 hpf, the margin of Pax6-positive lens epithelium (red) is associated with the retinal CMZ (light blue). Lens fiber cells are defined as Pax6-negative lens cells (green). The interface between the anterior and posterior half of the lens sphere is indicated by a dotted line. Index of nucleus flatness (Inf) is calculated as the ratio of the maximum (Lmax) to the minimum (Lmin) nuclear thickness. Inf=1.0-2.0, dark blue; 2.0-3.0, light blue; 3.0-4.0, green; 4.0-5.0, yellow; more than 5.0, red. (B-I) EdU incorporation (green) and DAPI labeling (red) of wild-type (B-E) and the vov mutant (F-I) zebrafish lens. Asterisk shows non-specific EdU signals (I). (J,K) Nuclear staining (upper panels) of wild-type (J) and the vov mutant (K) lens with DAPI. Inf is plotted to individual nuclei of lens fiber cells on the lower row of images. The degree of flatness is indicated by the height of bars and their color (see A). Dotted lines indicate the interface between the anterior and posterior halves. (L) The percentage of EdU-positive cells relative to the total number of lens epithelial cells in wild type (light blue bars) and the vov mutant (red bars). (M) The number of lens fiber cell nuclei in the wild type (light blue bars) and vov mutant (red bars). (N) Index of nuclear anterior localization (Ina) of the wild type and vov mutant. (O) The Inf of individual lens fiber cell nuclei in the wild type and vov mutant. Scale bars: 50 µm. PHENOTYPE:
|
|
Morphogenesis of lens fiber cells is affected in the vov mutant. (A-D2) Confocal images of the lens of 50 hpf wild type (A) and vov mutant (B) and 72 hpf wild type (C) and vov mutant (D) zebrafish expressing the SAGFF168A; UAS:EGFP transgenes. Individual lens fiber cells are indicated by pseudocolors (A2-D2). In the vov mutant, GFP-expressing cells do not maintain a fiber-like structure and appear to undergo degradation (D, arrowheads). (E,F) Anterior view of 72 hpf wild-type (E) and the vov mutant (F) lens labeled with Rhodamine-conjugated phalloidin (red) and Sytox Green (green). (G-N′) Transplantation of wild-type (G-I) and vov mutant (J-N) donor cells into wild-type recipient lens. Labeling is with biotin-dextran (green) (G,J), TOPRO3 (gray) (H,K) and phalloidin (red) (I,L). (M,N) High magnification of the boxed regions shown in J. Wild-type donor cells transplanted into wild-type recipient lens elongate to be very thin, resulting in weak signals (arrows, G). Nuclei of vov mutant donor lens fiber cells were frequently positioned in the anterior lens fiber region (yellow arrowheads, K,M,M2) and F-actin density is lower in the vov mutant (white arrows, L,N,N2). Scale bars: 50 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
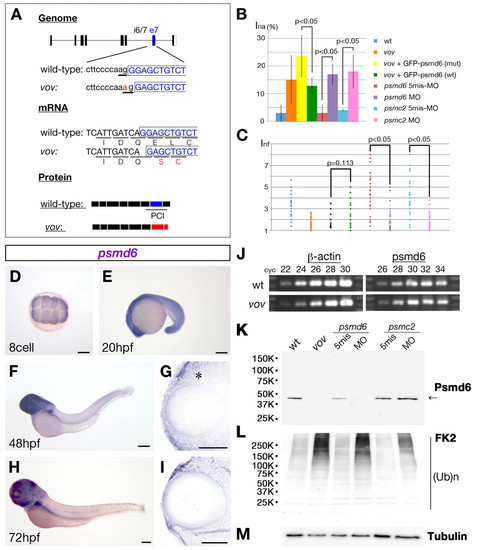
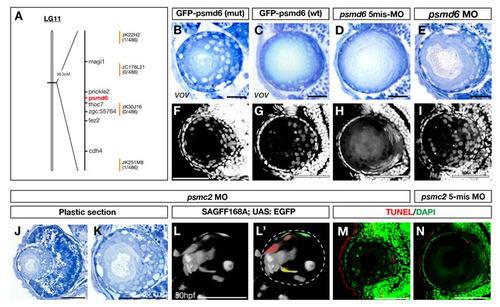
The vov mutant gene encodes Psmd6. (A) Nucleotide substitution from G to A occurs in a splicing acceptor site of the intron6/7-exon7 boundary (top, underlined), causing a one-base deletion in the psmd6 mRNA that results in a frameshift within the coding region (middle panel) and loss of the PCI domain (bottom panel). (B,C) Ina (B) and Inf (C) of wild type, vov mutant, vov mutant injected with GFP-tagged psmd6 RNA carrying the vov mutation, vov mutant injected with wild-type GFP-tagged psmd6 RNA, psmd6 and psmc2 morphants and their five-mismatch morphants. Ina is lower and the maximum value of Inf is higher in the vov mutant injected with wild-type GFP-tagged-psmd6 RNA (green) than in the vov mutant injected with GFP-tagged-psmd6 RNA carrying the vov mutation (yellow), suggesting that wild-type psmd6 RNA injection rescues the vov-mediated defects in nuclear position and shape of lens fiber cells. Ina is higher and the maximum value of Inf is lower in psmd6 (purple) and pmsc2 morphants (pink) than in their five-mismatch morphants (brown and light blue), suggesting that these MOs induce the vov-like lens defects. (D-I) Expression of zebrafish psmd6 mRNA at the eight-cell stage (D) and at 20 (E), 48 (F,G) and 72 hpf (H,I). (G,I) Plastic sections of eyes. Asterisk indicates psmd6 expression in the retinal CMZ. (J) Semi-quantitative PCR of wild type and vov mutant at 72 hpf using psmd6 and β-actin gene primers. The number of PCR cycles is indicated above each lanes. (K-M) Western blotting of 72 hpf wild type, vov mutant, psmd6 and psmc2 morphants and their five-mismatch morphants with anti-zebrafish Psmd6 (K), anti-polyubiquitylated protein (FK2) (L) and anti-acetylated α-tubulin (M, a loading control). The level of Psmd6 (arrow) is reduced in the vov mutant and psmd6 morphant, but is unaffected in the psmc2 morphant (K). The levels of polyubiquitylated proteins increase in the vov mutant, psmd6 and psmc2 morphant (K). Scale bars: 250 µm in D-F,H; 50 µm in G,I. EXPRESSION / LABELING:
PHENOTYPE:
|
|
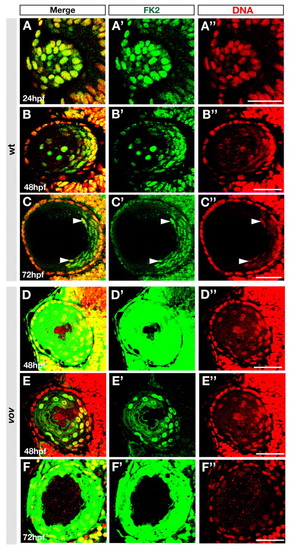
Polyubiquitylated proteins are localized in lens fiber cell nuclei. (A-F) Labeling of wild-type (A-C) and vov mutant (D-F) zebrafish lens of the indicated stages with anti-polyubiquitylated protein antibody (FK2, green) and TOPRO3 (red). Arrowheads indicate signals in nuclei of differentiating lens fiber cells at 72 hpf (C). Scanning conditions of images in D,F were the same as those for B,C. (E) The same image as D but with reduced sensitivity of fluorescence detection. (A′-F′,A"-F") The same images as A-F but shown through a green (FK2) or red (TOPRO3) channel. Scale bars: 50 µm. PHENOTYPE:
|
|
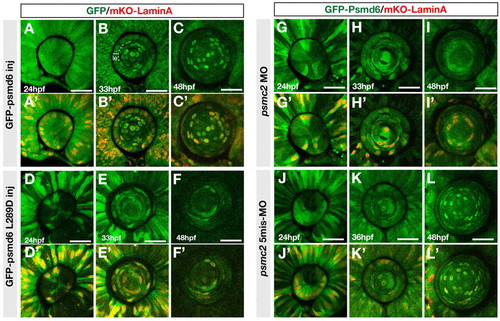
Localization of GFP-tagged Psmd6 in lens fiber cell nuclei. (A-F′) Localization of GFP-tagged Psmd6 (green) (A-C) and GFP-tagged Psmd6 carrying the L289D mutation (green) (D-F) in wild-type zebrafish lens of the indicated stages. Nuclear membranes are visualized with mKO-Lamin A (red) (A′-F′). le, lens epithelial region. (G-L′) Localization of GFP-tagged Psmd6 in psmc2 morphant (G-I) and psmc2 five-mismatch morphant (J-L) lens. Nuclear membranes are visualized with mKO-Lamin A (red) (G′-L′). All panels are anterior views of the lens sphere. Scale bars: 50 µm. |
|
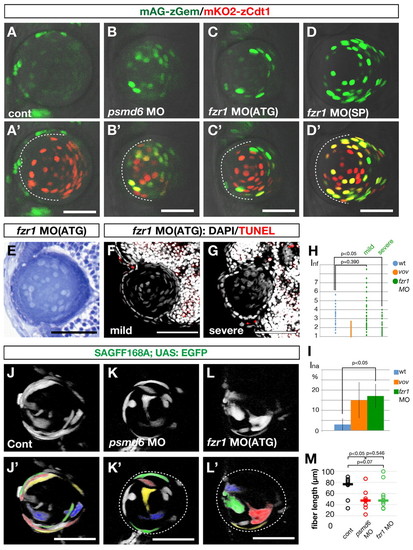
APC/C is required for lens fiber differentiation. (A-D′) Confocal images of 48 hpf wild type (A′), psmd6 morphant (B′), fzr1 ATG-morphant (C′) and fzr1 splicing-morphant (D′) zebrafish lens in the Cecyil transgenic background. (A-D) Green channel (mAG-zGem) only. Dashed lines indicate the interface between the lens epithelium and lens fiber core. mAG-zGem and mKO2-zCdt1 (red) expression overlap in the lens fiber cells of psmd6 and fzr1 morphants. mAG-zGem (green) expression is absent in the epithelium of psmd6 (B′) and fzr1 (C′,D′) morphant lens. The reduction of mAG-zGem seems to correlate with the absence of BrdU/EdU incorporation (Fig. 2L and see Fig. S9 in the supplementary material). (E) Lens of 72 hpf fzr1 morphant. (F,G) TUNEL (red) and DAPI (gray) labeling of 72 hpf lens of mild (F) and severe (G) fzr1 morphants. (H) Inf of wild type, vov mutant, and mild and severe fzr1 morphants at 72 hpf. (I) Ina for wild type, vov mutant and fzr1 morphant at 72 hpf. (J-L′) Confocal images of the lens of 48 hpf wild type (J), psmd6 morphant (K) and fzr1 morphant (L) expressing the SAGFF168A; UAS:EGFP transgenes. Individual lens fiber cells are indicated by pseudocolors (J′-L′). (M) Individual lens fiber lengths of wild type, psmd6 morphant and fzr1 morphant. The average fiber length (horizontal bars) is 50% shorter in both morphants than in the wild type. Scale bars: 50 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
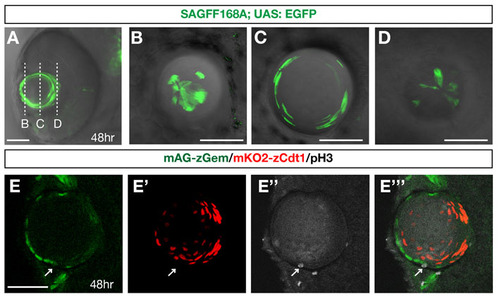
Zebrafish transgenic strains, SAGFF168A and Cecyil. (A) Ventral view of a zebrafish eye with the SAGFF168A; UAS:EGFP transgene at 48 hpf. GFP (green) is expressed in the lens. (B-D) Scanning of planes at the anterior-most surface (B), near the equator (C) and the posterior-most region of the lens sphere of 48 hpf SAGFF168A; UAS:EGFP transgenic embryo. GFP-positive lens fibers elongate to become very flat at the equatorial region (C) and extend their termini towards anterior and posterior poles, where tips of lens fiber processes converge (B,D). (E-E′′′) Confocal images of lens of Cecyil transgenic embryos with mAG-zGem (green), mKO2-zCdt1 (red) and anti-phosphorylated histone H3 antibody (gray, E′′). In the Cecyil transgenic line, an N-terminal 190 amino acid domain of zebrafish Cdt1 tagged with mKO2 (mKO2-zCdt1) (red, E′) and an N-terminal 100 amino acid domain of zebrafish Geminin tagged with mAG (mAG-zGem) (green, E) are expressed under the control of the EF1a promoter. mAG-zGem and mKO2-zCdt1 are expressed in lens epithelial cells and in postmitotic lens fiber differentiating cells, respectively. The anterior lens epithelial cells undergoing M phase do not express mAG-zGem or mKO2-zCdt1 (arrow). E′′′ is a merged image of E, E′ and E′′. Scale bars: 50 µm. |
|
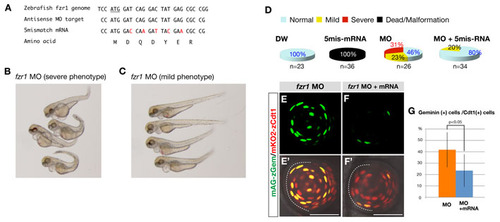
Sequences and specificity of fzr1-MO. (A) Alignment of the target sequences of antisense fzr1-MO and the corresponding sequences of five-mismatch fzr1 mRNA. Mismatched nucleotides are indicated in red. The deduced amino acid sequence of five-mismatch fzr1 mRNA is identical to that of wild-type fzr1 mRNA. The codon of the initial methionine is underlined. (B,C) 72 hpf embryos injected with fzr1-MO. These embryos were classified into two groups showing morphologically severe (B) and mild (C) defects. (D) Pie charts for severities of morphological defects in wild-type embryos injected with distilled water (DW), five-mismatch fzr1 RNA (50 µg/ml), fzr1-MO (0.25 mM) and both MO and mRNA. The injection of wild-type fzr1 RNA (data not shown) or five-mismatch fzr1 RNA into wild-type eggs causes severe malformation and embryonic death during gastrulation, suggesting that a high level of fzr1 RNA is toxic. However, the injection of five-mismatch fzr1 RNA decreases the morphological defects in fzr1 morphants as well as the rate of embryonic death, suggesting a counterbalance between five-mismatch fzr1 RNA and fzr1-MO. (E-F′) Ventral views of the Cecyil transgenic embryos injected with fzr1-MO (E′) and with both fzr1-MO and five-mismatch fzr1 mRNA (F′). (E,F) Green channel (mAG-Gem) only. Injection of fzr1-MO induces the colocalization of mAG-zGem (green) and mKO2-zCdt1 (red) (E,E′), suggesting that APC/C-mediated degradation of mAG-Gem is compromised. By contrast, the injection of both fzr1-MO and five-mismatch fzr1 mRNA inhibits the accumulation of mAG-Gem in lens fiber cells (F,F′), suggesting that five-mismatch fzr1 RNA inhibits defects in the degradation of mAG-Gem in the fzr1 morphant. (G) The percentage of mAG-zGem-positive cells relative to the total number of mKO2-zCdt1-positive lens fiber cells. The fraction of lens fiber cells expressing both mAG-zGem and mKO2-zCdt1 is significantly decreased in the fzr1 morphant injected with five-mismatch fzr1 RNA (P<0.05). The P-value was determined by Student′s t-test using n=9 for the fzr1 morphant and n=10 for the fzr1 morphant injected with five-mismatch fzr1 RNA. Scale bars: 50 µm. |
|
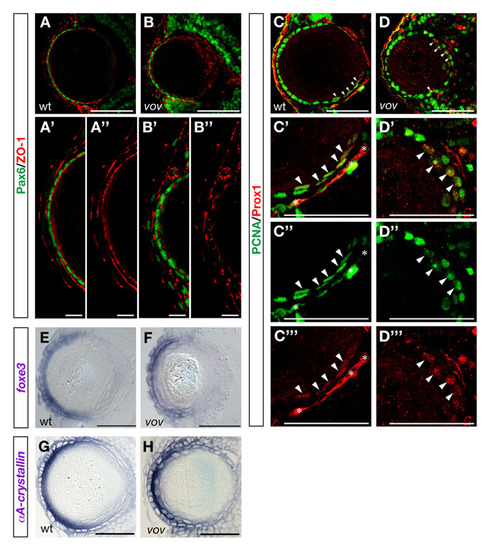
Expression of Pax6, ZO-1, Pcna, Prox1, foxe3 and αA-crystallin mRNA in wild-type and vov mutant embryos. (A,B) Expression of Pax6 (green) and ZO-1 (red) in wild-type (A) and vov mutant (B) lens at 72 hpf. (A′,B′) High magnification of anterior lens epithelium in A and B. (A′′,′′2) The same images of A and B shown through a red channel (ZO-1). In the vov mutant, ZO-1 signals underneath anterior lens epithelium are weaker than in wild type and sometimes discontinuous (B′′). (C,D) Expression of Pcna (green) and Prox1 (red) in wild-type (C) and vov mutant (D) lens at 72 hpf. In wild type, Pcna (green) is expressed strongly in the anterior lens epithelium and also weakly in four or five cells that pass over the region adjacent to the retinal CMZ (arrowheads, C); the latter are labeled with Prox1 (red). In the vov mutant, Pcna (green) is not only expressed in the anterior lens epithelium but also in cells located at the posterior edge of the lens sphere. Some Pcna-expressing cells strongly express Prox1 (red) at the posterior lens edge in the vov mutant (arrowheads, D). (C′) High magnification of the posterior lens region of C. (C′′,C′′′) The same image as C′ through a green (Pcna) or red (Prox1) channel. The Pcna and Prox1 double-positive cells seem to be early differentiating lens fiber cells. Asterisks indicate cells of intraocular blood vessels. (D′) High magnification of the posterior lens region in D. (D′′,D′′′) The same image as D2 through a green (Pcna) or red (Prox1) channel. Most posterior Pcna-positive cells are labeled with Prox1 (red, arrowheads), suggesting that posterior-positioned Pcna cells in the vov mutant are early differentiating lens fiber cells. (E,F) foxe3 mRNA expression of wild-type (E) and the vov mutant (F) lens at 72 hpf. foxe3 mRNA is expressed in the lens epithelium in wild type and the vov mutant. (G,H) Expression of crystallin αA mRNA in 72 hpf wild-type (N) and vov mutant (O) lens. crystallin αA mRNA is strongly expressed in differentiating lens fiber cells in wild type and the vov mutant. Scale bars: 50 µm, except 10 µm in A′,A′′,B′,B′′. |
|
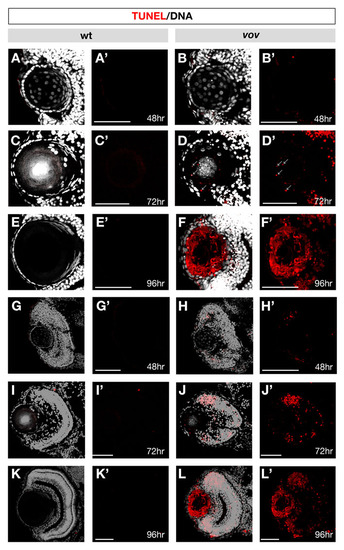
TUNEL in wild-type and the vov mutant lens and retina. (A-L) TUNEL (red) and DAPI (gray) staining of wild-type lens (A,C,E) and retina (G,I,K) and vov mutant lens (B,D,F) and retina (H,J,L) at 48 hpf (A,B,G,H), 72 hpf (C,D,I,J) and 96 hpf (E,F,K,L). (A′-L′) The same images as A-L shown through only a red channel (TUNEL). No TUNEL signal could be detected in the wild-type lens and retina at 48, 72 and 96 hpf and vov mutant lens at 48 hpf. However, in the vov mutant, small dotted signals are observed at 72 hpf, and most of the lens fiber cells surrounding a small lens fiber core are TUNEL positive, whereas the lens epithelium is TUNEL negative, suggesting that a majority of lens fiber cells undergo apoptosis-like degradation in the vov mutant at 96 hpf, except for a small central lens fiber core. In the vov mutant retina, TUNEL signals are observed in the neurogenic region near retinal CMZ at 48 hpf (H,H′). TUNEL signals increase at 72 hpf (J,J′) and 96 hpf (L,L′). Scale bars: 50 µm. |
|
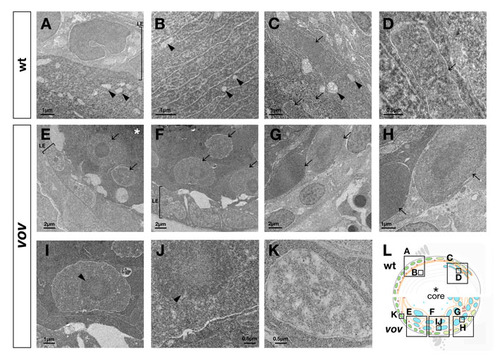
Ultrastructure of the vov mutant lens. (A-D) Transmission electron microscopy (TEM) images of wild-type lens at 72 hpf. No nuclei of lens fiber cells were observed underneath the anterior lens epithelium (A). Arrowheads indicate subcellular structures similar to mitochondria in the lens fiber region underneath the anterior lens epithelium (arrowheads, A-C). In the posterior region of the lens sphere, flat nuclei of lens fiber cells are stacked regularly (arrows, C,D). Nuclei of lens fiber cells were electron-densely stained in patches (D). (E-K) TEM images of the vov mutant lens at 72 hpf. Large nuclei are observed underneath the anterior epithelium (arrows, E), near the equator (arrows, F) and in the posterior region of the lens sphere (arrows, G,H). Unlike in the wild type, most of the lens fiber nuclei were stained uniformly, and also contained a dark-stained circular region inside (arrowhead, I,J). In contrast to lens fiber cells, nuclei of lens epithelium were electron-densely stained in patches (K). (L) Schematic of wild-type and vov mutant lens. Boxes indicate areas shown in A-K. Asterisks indicate the lens fiber core, which is darkly stained in TEM analyses (A,C). LE, lens epithelium. |
|
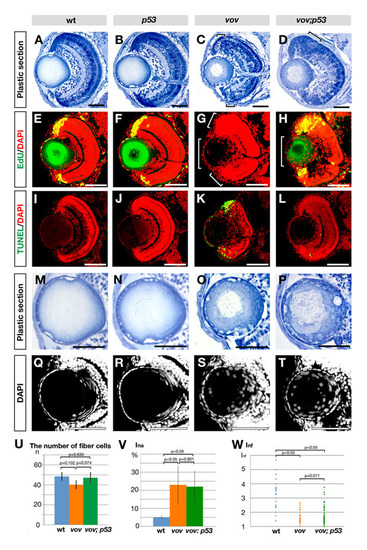
Retinal and lens phenotypes of zebrafish vov; p53 double mutant. (A-D) Plastic sections of wild-type (A), p53 (B), vov (C) and vov; p53 double mutant (D) eyes at 72 hpf. Apoptosis near the CMZ in the vov mutant (C, small square brackets) is inhibited in the vov; p53 double mutant (D). These surviving retinal cells do not laminate in the vov; p53 double mutant (D, large square bracket), suggesting that UPS is required for neuronal differentiation of retinal cells in a p53-independent manner. (E-H) EdU incorporation in wild-type (E), p53 (F), vov (G) and vov; p53 double mutant (H) lens at 72 hpf. Nuclei were counterstained with DAPI (red). EdU incorporation (green) does not occur in the retinal CMZ (small square bracket) and lens epithelium (large square bracket) in the vov mutant (G). In the vov; p53 double mutant, EdU incorporation is restored in the retinal CMZ, but the EdU-positive CMZ region is larger than in wild types and p53 mutants (H), suggesting that CMZ cells fail to exit from the cell cycle. EdU incorporation did not recover in the lens epithelium of the vov; p53 double mutant (H, large square bracket). (I-L) TUNEL staining (green) of wild-type (I), p53 (J), vov (K) and vov; p53 double mutant (L) lens at 72 hpf. Nuclei were counterstained with DAPI (red). TUNEL is detected in the neurogenic region near retinal CMZ in the vov mutant (K), whereas retinal apoptosis is suppressed in the vov; p53 double mutant (L). (M-P) Plastic sections of wild-type (M), p53 (N), vov (O) and vov; p53 double mutant (P) lens at 72 hpf. The vov; p53 double mutant shows defects in nuclear shape and position of lens fiber cells (P), which are similar to those of the vov mutant (O). (Q-T) Nuclear staining of wild-type (Q), p53 (R), vov (S) and vov; p53 double mutant (T) lens with DAPI at 72 hpf. (U) The number of lens fiber cell nuclei in the wild-type, vov, and vov; p53 double mutant at 72 hpf. The number of lens fiber cell nuclei in the vov; p53 double mutant is similar to that of wild type. (V) Ina of the vov; p53 double mutant is higher than that of wild type and similar to that of the vov mutant. The vov defects in nuclear position of lens fiber cells are not inhibited by the p53 mutation. (W) Inf of the wild type, vov, and vov; p53 double mutant. Like the vov mutant, the maximum Inf value of the vov; p53 double mutant is lower than that of the wild type, suggesting that the vov defects in nuclear shape of lens fiber cells are not inhibited by the p53 mutation. Student′s t-test (U,V,W). Scale bars: 50 μm. |
|
Lens phenotypes of vov mutants injected with GFP-tagged wild-type and vov mutant form psmd6 RNA and of psmd6 and psmc2 morphants. (A) The vov mutation is mapped in the genomic region between two polymorphic markers, zK22H2 and zK251M8, on zebrafish chromosome 11 (Vega Ver.32). Seven genes are annotated within this genomic region. (B,C) 72 hpf lens of vov mutant embryos injected with RNA encoding GFP-tagged Psmd6 carrying the vov mutation (B) and GFP-tagged wild-type Psmd6 (C). The introduction of wild-type psmd6 RNA inhibits the lens defects in the vov mutant, although some lens fiber nuclei are round in shape (C). (D,E) 72 hpf lens of the psmd6 five-mismatch morphant (D) and psmd6 morphant (E). The psmd6 morphant shows lens defects similar to the vov mutant. (F,G) DAPI labeling of 72 hpf lens of the vov mutant embryos injected with mRNA encoding GFP-tagged Psmd6 carrying the vov mutation (F) and GFP-tagged wild-type Psmd6 (G). The number of lens fiber nuclei positioned in the anterior half of the lens sphere is lower in vov mutant injected with wild-type psmd6 RNA than that of the vov-mutant form of psmd6 RNA. (H,I) DAPI labeling of 72 hpf lens of psmd6 five-mismatch morphant (H) and psmd6 morphant (I). Nuclei are swollen and retained in the anterior half of the lens sphere in the psmd6 morphant. (J,K) Plastic sections of 72 hpf psmc2 morphant eyes (J) and lens (K). Like the vov mutant, retinal apoptosis occurs (J) and round lens cell nuclei surround a small lens fiber core (K) in the psmc2 morphant lens. (L,L′) Confocal image of 50 hpf psmc2 morphant under the SAGFF168A; UAS:EGFP transgene. Lens fiber cells fail to fully elongate (pseudocolors in L′). (M,N) TUNEL of 72 hpf lens of psmc2 morphant (M) and psmc2 five-mismatch morphant (N). A few TUNEL signals (red) were detected in both cases. Nuclei were counterstained by DAPI (green). Scale bars: 50 μm. |
|
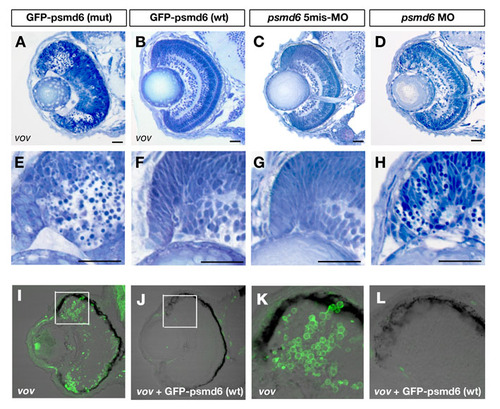
Retinal phenotypes of vov mutants injected with GFP-tagged wild-type and vov mutant form psmd6 RNA and of psmd6 morphant. (A,B) Plastic sections of vov mutants injected with GFP-tagged wild-type (A) and vov mutant form psmd6 RNA (B) at 72 hpf. (E,F) Higher magnification of retinal CMZ of A,B. Pyknotic nuclei are observed in the neurogenic region of retinal CMZ of the vov mutant injected with GFP-tagged psmd6 mutant form RNA. By contrast, apoptosis in retinal CMZ is suppressed and retinal lamination is normal in the vov mutant injected with GFP-tagged wild-type psmd6 RNA. (C,D) Plastic sections of retinas of psmd6 five-mismatch morphant (C) and psmd6 morphant (D) at 72 hpf. (G,H) Higher magnification of retinal CMZ of C,D. Apoptosis is observed in retinal CMZ of psmd6 morphant (D), but not in psmd6 five-mismatch morphant (C). (I-L) TUNEL of vov mutant (I,K) and vov mutant injected with wild-type psmd6 RNA (J,L). (K,L) High-magnification images of boxed regions in I,J. The number of TUNEL-positive cells in the retinal CMZ is significantly reduced in the vov mutant injected with psmd6 RNA as compared with the vov mutant. Scale bars: 50 μm. |
|
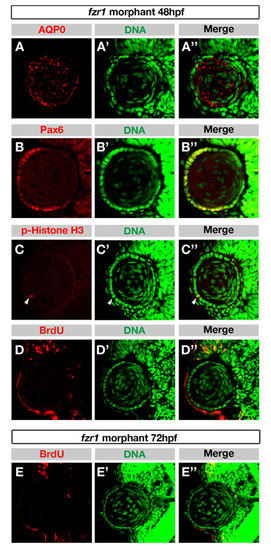
In the fzr1 morphant, proliferation of lens epithelial cells is affected, but specification of lens epithelium and lens fiber cells occur at 48 hpf. (A-C′′) Labeling of 48 hpf fzr1 morphant lens with anti-AQP0 (A), anti-Pax6 (B) and anti-pH3 (C) antibodies. Nuclei were labeled with DAPI (A′-C′); merge (A′′-C′′) AQP0 and Pax6 were expressed in lens fiber cells and lens epithelial cells, respectively. pH3-positive epithelial cells were not increased in number (arrowhead in C-C′′), suggesting that cell cycle arrest in M phase is unlikely in the fzr1 morphant. (D-E′′) BrdU incorporation of fzr1 morphant at 48 hpf (D) and 72 hpf (E). Nuclei are labeled with DAPI (D′,E′); merge (D′′,E′′). No BrdU incorporation was detected in lens epithelium in fzr1 morphant at both stages, suggesting that cell proliferation is affected in the fzr1 morphant. |