Fig. 6
|
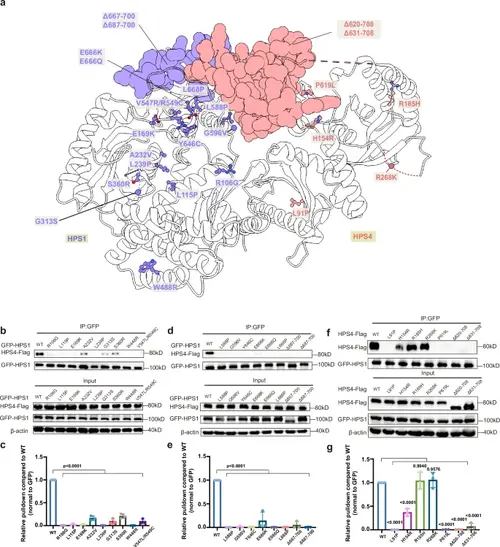
Structural mapping of the disease-related mutations and their effects on BLOC-3 assembly.a Structural mapping of the disease-associated mutations of BLOC-3. b–e Coimmunoprecipitation assays were used to assess the effects of the disease-associated mutations on the formation of BLOC-3. HPS1 mutation variants were cotransfected with Flag-tagged HPS4 in HEK293F cells, the cells were lysed and coimmunoprecipitated with anti-GFP affinity gel, and the purified samples and input samples were immunoblotted. Antibodies: anti-Flag (HPS4), anti-GFP (HPS1) and anti-β-Actin. (c) and (e) presented the quantification of the results presented in (b) and (d), respectively. Statistical differences were determined by One-way ANOVA. Error bars represent the standard deviation of three independent biological repeats. n = 3. f, g HPS4 mutation variants were cotransfected with GFP-tagged HPS1 in HEK293F cells, the cells were lysed and coimmunoprecipitated with anti-GFP affinity gel, and the purified samples and input samples were immunoblotted. Antibodies: anti-Flag (HPS4), anti-GFP (HPS1) and anti-β-Actin. (g) presented the quantification of the results presented in (f). Statistical differences were determined by One-way ANOVA. Error bars represent the standard deviation of three independent biological repeats. n = 3. |