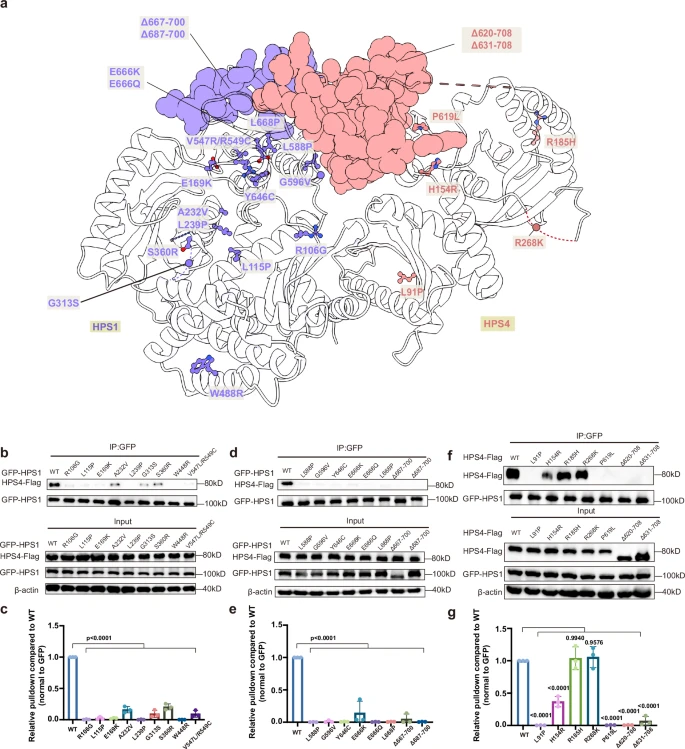
Fig. 6 Structural mapping of the disease-related mutations and their effects on BLOC-3 assembly.a Structural mapping of the disease-associated mutations of BLOC-3. b–e Coimmunoprecipitation assays were used to assess the effects of the disease-associated mutations on the formation of BLOC-3. HPS1 mutation variants were cotransfected with Flag-tagged HPS4 in HEK293F cells, the cells were lysed and coimmunoprecipitated with anti-GFP affinity gel, and the purified samples and input samples were immunoblotted. Antibodies: anti-Flag (HPS4), anti-GFP (HPS1) and anti-β-Actin. (c) and (e) presented the quantification of the results presented in (b) and (d), respectively. Statistical differences were determined by One-way ANOVA. Error bars represent the standard deviation of three independent biological repeats. n = 3. f, g HPS4 mutation variants were cotransfected with GFP-tagged HPS1 in HEK293F cells, the cells were lysed and coimmunoprecipitated with anti-GFP affinity gel, and the purified samples and input samples were immunoblotted. Antibodies: anti-Flag (HPS4), anti-GFP (HPS1) and anti-β-Actin. (g) presented the quantification of the results presented in (f). Statistical differences were determined by One-way ANOVA. Error bars represent the standard deviation of three independent biological repeats. n = 3.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Commun.