FIGURE 2
- ID
- ZDB-FIG-241219-94
- Publication
- Zheng et al., 2024 - Low-input CUT&Tag for efficient epigenomic profiling of zebrafish stage I oocytes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
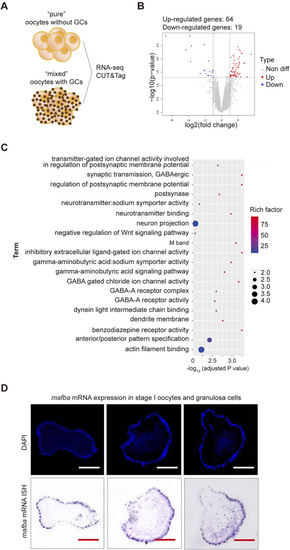
RNA sequencing analysis of differentially expressed genes (DEGs) between stage I oocytes with or without granulosa cells. |