- Title
-
Low-input CUT&Tag for efficient epigenomic profiling of zebrafish stage I oocytes
- Authors
- Zheng, Q., Wu, X., Li, X., Mo, X., Xiang, B., Chen, J.
- Source
- Full text @ Front Cell Dev Biol
|
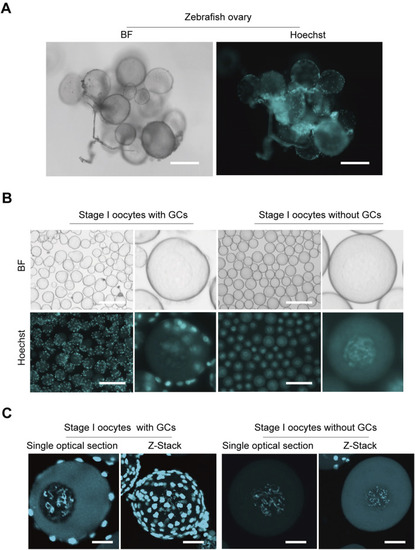
Isolation zebrafish stage I oocytes without granulosa cells. |
|
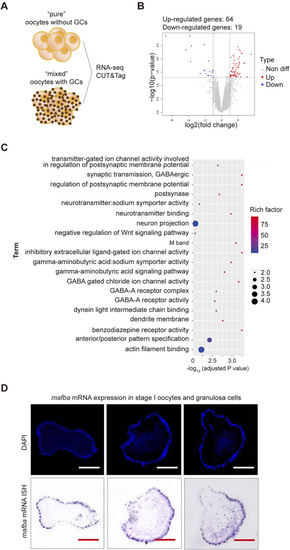
RNA sequencing analysis of differentially expressed genes (DEGs) between stage I oocytes with or without granulosa cells. |
|
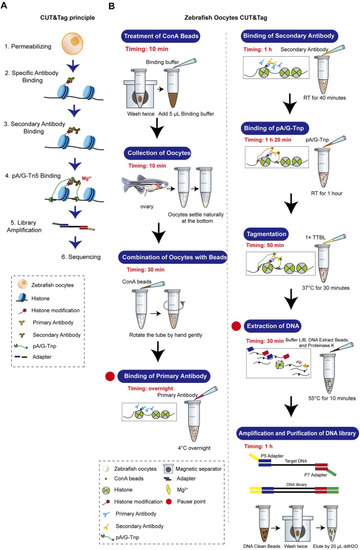
Zebrafish oocyte CUT&Tag principle and workflow. |
|
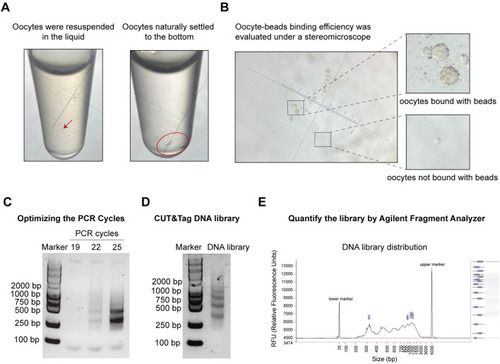
Pre-sequencing quality control of oocyte CUT&Tag. |
|
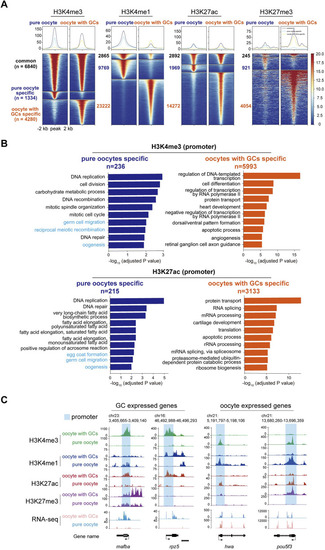
CUT&Tag analysis of histone modifications between stage I oocytes with or without granulosa cells. |
|
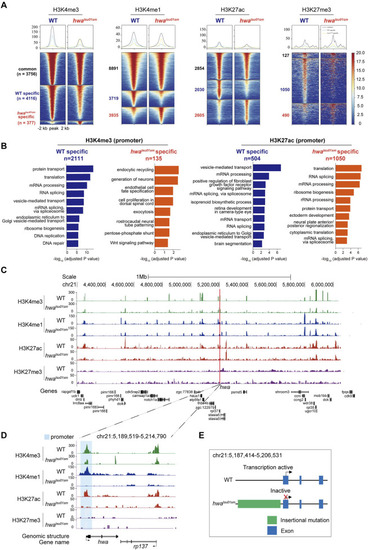
Distinct histone modifications around |