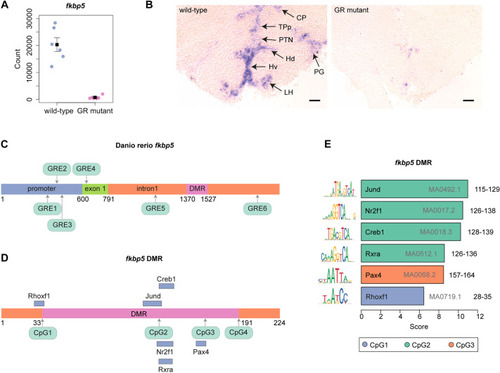
fkbp5 is differentially expressed and differentially methylated in wild-type and GR-mutant zebrafish brains. (A) GR function is required for fkbp5 expression in the adult brain. Normalised transcript-count data for fkbp5 obtained from RNA-Seq analysis of wild-type and GR-mutant fish (both n=6) adult brains (P=3.45×10−25). (B) GR function is required for fkbp5 expression in the adult brain. In situ hybridisation analysis of fkbp5 transcripts in transverse sections of adult zebrafish brain, cross section 149 (Wulliman et al., 1996) from wild-type (left panel) and GR-mutant (right panel) adult males, at a rostro-caudal position through the thalamus (CP, central posterior thalamic nucleus), the posterior tuberculum (TPp, periventricular nucleus of posterior tuberculum; PTN, posterior tuberal nucleus; PG, preglomerular nucleus) and the hypothalamus (Hd and Hv, dorsal and ventral zone of periventricular hypothalamus, respectively; LH, lateral hypothalamic nucleus). Results show fkbp5 expression in wild-type brain that is almost completely extinguished in the GR-mutant brain. Wild-type and GR-mutant (both n=6) fish, across three independent experiments. Scale bars: 100 μm. (C) Schematic of fkbp5, showing promoter region, exon 1 and intron1. Six distinct GR-binding sites (GREs) within the promoter region (GRE1, GRE2, GRE3), exon 1 (GRE4) and intron 1 (GRE5, GRE6) are shown in green. (D) Schematic of the differentially methylated region (DMR; pink) within fkbp5, surrounded either side by two 33-bp-long flanking sequences (orange). Four cytosine and guanine dinucleotides separated by a phosphate, i.e. CpG1, CpG2, CpG3, CpG4, are shown in green (see Fig. 2A for chromosome 6 nucleotide coordinates). Binding sites for transcription factors Rhoxf1, Jund, Nr2f1, Rxra, Creb1 and Pax4 overlap with CpGs, and are shown as blue bars. (E) Bar graph of data obtained by using the JASPAR database, indicating that CpG1 of the fkbp5 DMR is located within the binding motif for transcription factor RHOXF1, CpG2 is located within the binding motifs for transcription factors Creb1, Jund, Nr2f1 and Rxra, and CpG3 is located within the binding motif for transcription factor Pax4. Location of CpG4 is not located within a known transcription factor-binding motif. JASPAR matrix IDs are shown in grey within or to the right of each bar. DNA coordinates (nucleotide numbers) for each CpG within the fkbp5 intron 1 as depicted in panel D, are shown in black. The consensus sequence for the DNA-binding motif is shown to the left of each bar.
|