|
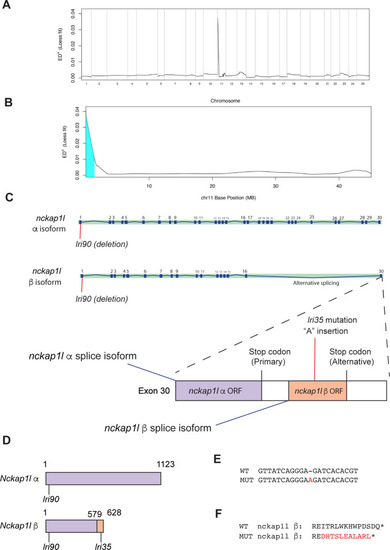
The <italic>lri35</italic> mutation disrupts the minor isoform of the <italic>nckap1l</italic> gene.(A) Mapping of the lri35 mutation. Whole genome-wide Loess fit curve for SNP allele frequency Euclidean distance computed by the MMAPPR algorithm. Chromosome 11 showed the highest score. (B) Loess fit curve for SNP allele frequency Euclidean distance within chromosome 11. The critical region was mapped to the distal tip of chromosome 11. (C) The nckap1l gene has two splice isoforms: the previously annotated major splice isoform (α isoform) and an unknown minor isoform (ß isoform). The lri35 mutation, which is a one-nucleotide insertion, is located in the last exon and induces a frameshift only in the ß isoform of the nckap1l gene. These data indicate that the minor isoform of the nckap1l gene is responsible for the phenotypes in lri35 mutant larvae. (D) Schematic of Nckap1l α and ß proteins. The entire Nckap1l α protein is recognized as the Nckap1l domain (purple box). Due to the alternative splicing of the nckap1l gene, Nckap1l ß shifted to the ß specific sequence (orange box) after the position 579. The lri90 mutation affects both Nckap1l α and ß, whereas the lri35 mutation affects Nckap1l ß only. (E) The lri35 mutation is an insertion mutation that inserts an additional adenine nucleotide into the nckap1l gene. (F) The lri35 mutation induces a frameshift and changes the last 13 amino acids of the ß isoform of the Nckap1l protein.
|