Fig. 3
- ID
- ZDB-FIG-210413-36
- Publication
- Klatt Shaw et al., 2021 - Localized EMT reprograms glial progenitors to promote spinal cord repair
- Other Figures
- All Figure Page
- Back to All Figure Page
|
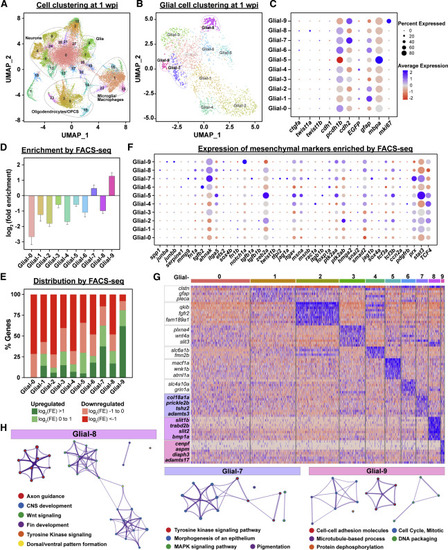
Single cell profiling of glial cells during SC regeneration (A) UMAP of snRNA-seq data from SC tissues at 1 wpi. (B) Sub-clustering and UMAP visualization of glial cell clusters at 1 wpi. Ten clusters of glial cells were identified and termed as Glial-0 to -9. (C) Expression of canonical markers in each glial cluster. Dot colors represent average expression levels from low (red) to high (blue). Dot sizes represent the percent of positive cells for each gene. (D) Enrichment of snRNA-seq-derived glial cell markers by FACS-seq. Glial cell markers were identified for each cluster by snRNA-seq analysis. Log2(fold enrichment) in sorted bridging glia relative to uninjured SCs were then then averaged for each cluster. (E) Distribution of snRNA-seq-derived glial cell markers by FACS-seq. Markers of clusters Glial-0 to -9 were categorized based on their enrichment in ctgfa+gfap+ cells. Upregulated genes were subdivided into log2(FE) > 1 (Green) and log2(FE) = 0 to 1 (Light green). Downregulated genes were subdivided into log2(FE) = −1 to 0 (light red) and log2(FE) < −1 (red). Percent genes within each category are shown. (F) Mesenchymal gene expression in glial clusters. A total of 39 mesenchymal markers that are enriched by FACS-seq are shown. Dot colors represent average expression levels from low (red) to high (blue). Dot sizes represent the percent of positive cells for each gene. (G) Heatmap of genes enriched in clusters Glial-0 to -9. Blue indicates enriched genes and red indicates depleted genes. (H) Gene ontology for markers of clusters Glial-7, -8, and -9. |
Reprinted from Developmental Cell, 56(5), Klatt Shaw, D., Saraswathy, V.M., Zhou, L., McAdow, A.R., Burris, B., Butka, E., Morris, S.A., Dietmann, S., Mokalled, M.H., Localized EMT reprograms glial progenitors to promote spinal cord repair, 613-626.e7, Copyright (2021) with permission from Elsevier. Full text @ Dev. Cell