Fig. S4
- ID
- ZDB-FIG-190814-6
- Publication
- Lin et al., 2019 - An Ectoderm-Derived Myeloid-like Cell Population Functions as Antigen Transporters for Langerhans Cells in Zebrafish Epidermis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
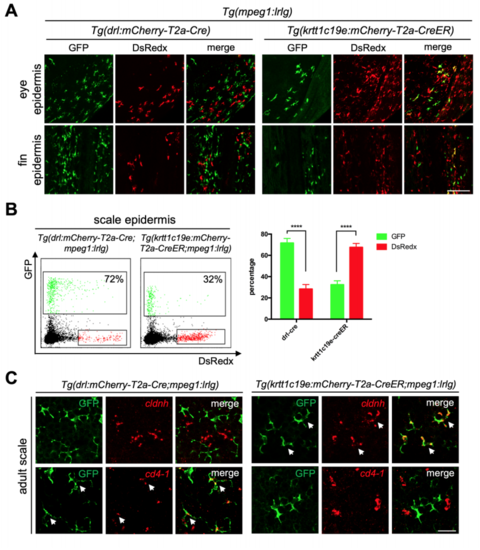
Ectodermal derived mpeg1+ populations only existed in epidermis. Related to Figure 3 (A) Confocal images of epidermis covered eyes and fins. GFP+ cells are cLCs and DsRedx+ cells are MLCs in Tg(drl:mCherry-T2a-Cre;mpeg1:loxP-DsRedxloxP-GFP). In Tg(krtt1c19e:mCherry-T2a-CreERT2;mpeg1:loxP-DsRedx-loxPGFP) fish (add 4-OHT at 1-5dpf), GFP+ cells are MLCs while DsRedx+ cells are cLCs. Scale bar, 100μm. (B) FACS analysis of mpeg1+ macrophage populations in scale epidermis. In Tg(drl:mCherry-T2a-Cre;mpeg1:loxP-DsRedx-loxP-GFP) fish, about 72% of total mpeg1+ cells are GFP+, while in Tg(krtt1c19e:mCherry-T2aCreERT2;mpeg1:loxP-DsRedx-loxP-GFP) (add 4-OHT at 1-5dpf) fish, only 32% of total mpeg1+ cells are GFP+. Quantification of GFP+ and DsRedx+ cells percentages to total mpeg1+ population in scale epidermis (each group, n=4). (C) In-situ hybridization of cldnh (red) and cd4-1 (red) in scales from Tg(krtt1c19e:mCherry-T2a-CreERT2;mpeg1:loxP-DsRedx-loxP-GFP) and Tg(drl:mCherry-T2a-Cre;mpeg1:loxP-DsRedx-loxP-GFP) fish, followed by antiGFP staining. White arrows indicate co-localization of cldnh, cd4-1 with GFP+ cells. Scale bar, 25μm. Data are represented as mean ± SD, ****P<0.0001. |
Reprinted from Developmental Cell, 49(4), Lin, X., Zhou, Q., Zhao, C., Lin, G., Xu, J., Wen, Z., An Ectoderm-Derived Myeloid-like Cell Population Functions as Antigen Transporters for Langerhans Cells in Zebrafish Epidermis, 605-617.e5, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell